The Optimization and Characterization of an RNA-Cleaving Fluorogenic DNAzyme Probe for MDA-MB-231 Cell Detection
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials and Reagents
2.2. Cell Lines and Cell Culture
2.3. Sample Preparation
2.4. Sequences Ligation Process
3. Results and Discussion
3.1. Optimization of the Reaction Buffer Conditions
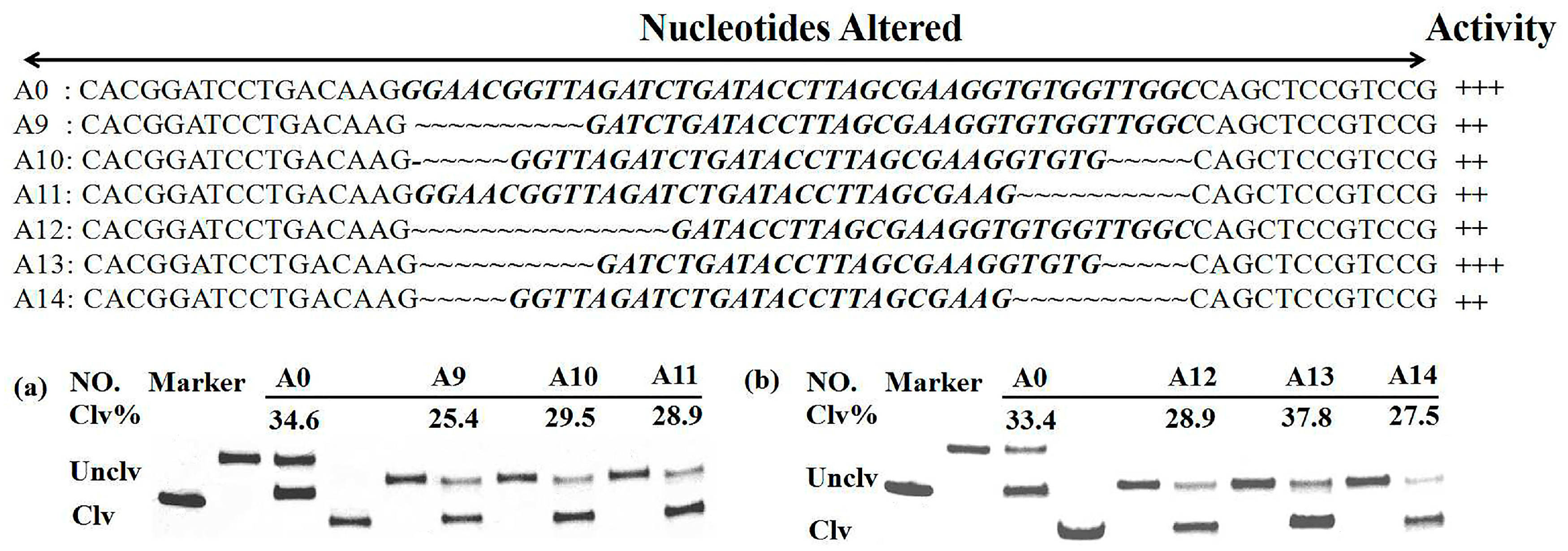
3.2. Sequence Minimization
3.3. Sensitivity of the Optimized Probe
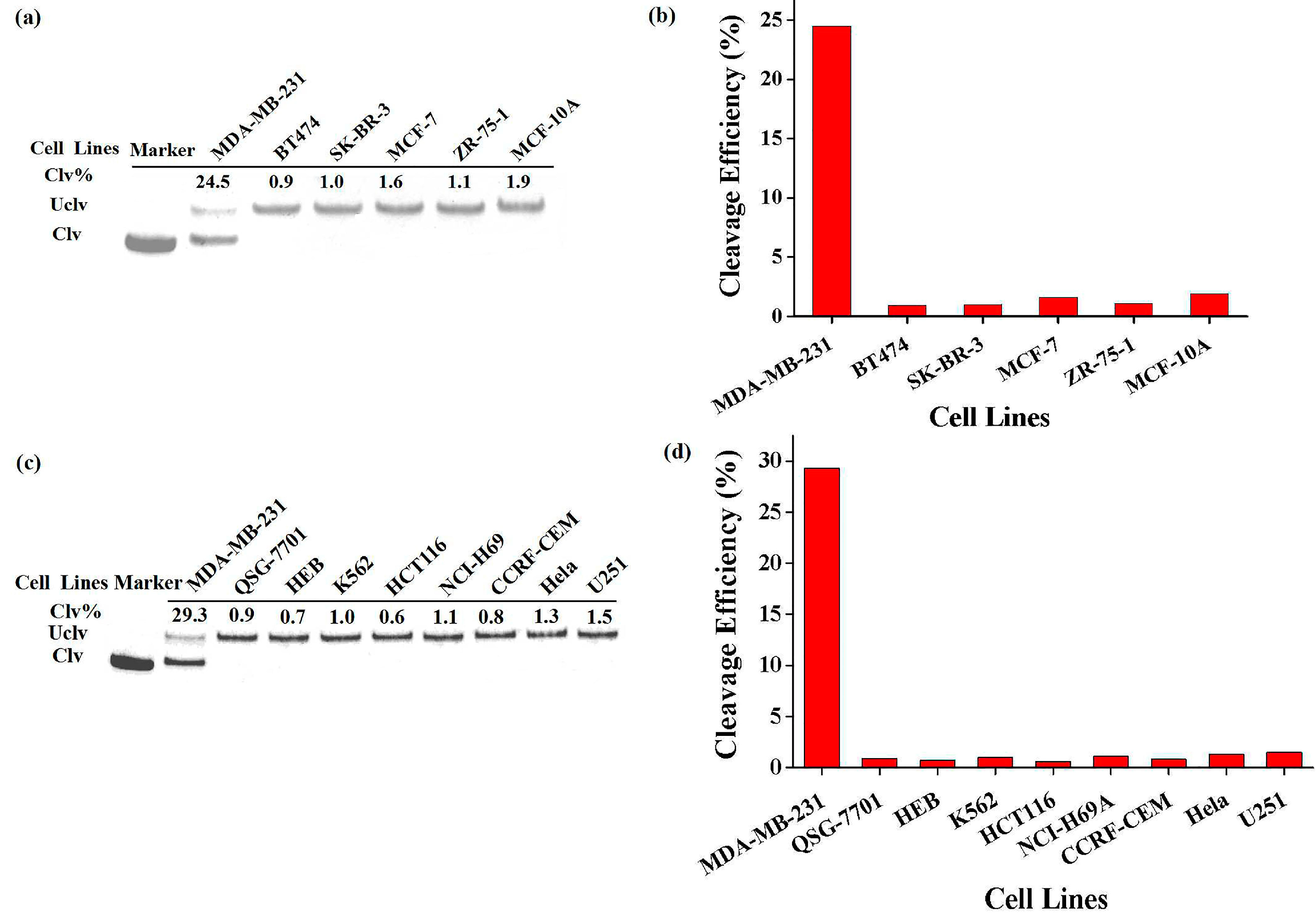
3.4. Specificity of the New Probe
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hippman, C.; Moshrefzadeh, A.; Lohn, Z.; Hodgson, Z.G.; Dewar, K.; Lam, M.; Albert, A.Y.K.; Kwong, J. Breast Cancer and Mammography Screening: Knowledge, Beliefs and Predictors for Asian Immigrant Women Attending a Specialized Clinic in British Columbia, Canada. J. Immigr. Minor. Health 2016, 18, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Machado, V.A.; Peixoto, D.; Queiroz, M.J.; Soares, R. Antiangiogenic 1-Aryl-3-[3-(thieno[3,2-b]pyridin-7-ylthio)phenyl]ureas Inhibit MCF-7 and MDA-MB-231 Human Breast Cancer Cell Lines Through PI3K/Akt and MAPK/Erk Pathways. J. Cell. Biochem. 2016, 117, 2791–2799. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Sun, G.Q.; Liu, F.; Lu, J.J.; Yu, J.H.; Song, X.R. An aptasensor for sensitive detection of human breast cancer cells by using porous GO/Au composites and porous PtFe alloy as effective sensing platform and signal amplification labels. Anal. Chim. Acta 2013, 798, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Fremgen, A.M.; Bland, K.I.; McGinnis, L.S.; Eyre, H.J.; McDonald, C.J.; Menck, H.R.; Murphy, G.P. Clinical highlights from the National Cancer Data Base, 1999. Cancer J. Clin. 1999, 49, 145–158. [Google Scholar] [CrossRef]
- Xie, Y.C.; Yin, T.; Wiegraebe, W.; He, X.C.; Miller, D.; Stark, D.; Perko, K.; Alexander, R.; Schwartz, J.; Grindley, J.; et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 2010, 457, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.; Chan, D.-Y.; Cheng, D.-C.; Ho, Y.-J.; Tsai, P.-P.; Shen, W.-C.; Chen, R.-F. Automated Feature Set Selection and Its Application to MCC Identification in Digital Mammograms for Breast Cancer Detection. Sensors 2013, 13, 4855–4875. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.F.; Guo, M.L.; Nie, Z.; Xiao, X.L.; Yao, S.Z. Aptamer-Based Electrochemical Sensor for Label-Free Recognition and Detection of Cancer Cells. Electroanalysis 2009, 21, 1321–1326. [Google Scholar] [CrossRef]
- Eltahir, E.M.; Mallinson, D.S.; Birnie, G.D.; Hagan, C.; George, W.D.; Purushotham, A.D. Putative markers for the detection of breast carcinoma cells in blood. Br. J. Cancer 1998, 77, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhu, X.L.; Zhang, D.M.; Huang, J.Y.; Li, G.X. An electrochemical method to detect folate receptor positive tumor cells. Electrochem. Commun. 2007, 9, 2547–2550. [Google Scholar] [CrossRef]
- Ehrhart, J.C.; Bennetau, B.; Renaud, L.; Madrange, J.P.; Thomas, L.; Morisot, J.; Brosseau, A.; Allano, S.; Tauc, P.; Tran, P.L. A new immunosensor for breast cancer cell detection using antibody-coated long alkylsilane self-assembled monolayers in a parallel plate flow chamber. Biosens. Bioelectron. 2008, 24, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Liu, C.; Tan, W.H. Aptamers generated by Cell SELEX for biomarker discovery. Biomark. Med. 2009, 3, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wu, Q.; Hamerlik, P.; Hitomi, M.; Sloan, A.E.; Barnett, G.H.; Weil, R.J.; Leahy, P.; Hjelmeland, A.B.; Rich, J.N. Aptamer Identification of Brain Tumor-Initiating Cells. Cancer Res. 2013, 73, 4923–4936. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.W.; Shangguan, D.; Wang, K.M.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W.H. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.H.; Meng, L.; Cao, Z.H.C.; Xiao, Z.Y.; Fang, X.H.; Li, Y.; Cardona, D.; Witek, R.P.; Liu, C.; Tan, W.H. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008, 80, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Herr, J.K.; Smith, J.E.; Medley, C.D.; Shangguan, D.H.; Tan, W.H. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal. Chem. 2006, 78, 2918–2924. [Google Scholar] [CrossRef] [PubMed]
- Medley, C.D.; Bamrungsap, S.; Tan, W.H.; Smith, J.E. Aptamer-Conjugated Nanoparticles for Cancer Cell Detection. Anal. Chem. 2011, 83, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.J.; Fang, X.H.; Tan, W.H. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.K.; Brewer, J.H.; Henry, J.M.; Harrington, D.J.; Carrigan, D.R. Human herpesvirus 6 and multiple sclerosis: Systemic active infections in patients with early disease. Clin. Infect Dis. 2000, 31, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Hamm, J.; Libri, D.; Tavitian, B.; de Franciscis, V. Nucleic acid aptamers in cancer medicine. FEBS Lett. 2002, 528, 12–16. [Google Scholar] [CrossRef]
- Gold, L. Oligonucleotides as Research, Diagnostic, and Therapeutic Agents. J. Biol. Chem. 1995, 270, 13581–13584. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, B.L.; Dong, S.J. Adaptive recognition of small molecules by nucleic acid aptamers through a label-free approach. Chem Eur J. 2007, 13, 6718–6723. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Zhang, W.Q.; Feng, Q.; Chang, D.R.; Tram, K.; Li, Y.F. In Vitro selection of RNA-cleaving DNAzymes for bacterial detection. Methods 2016, 106, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Aguirre, S.D.; Lazim, H.; Li, Y.F. Fluorogenic DNAzyme Probes as Bacterial Indicators. Angew. Chem. Int. Ed. 2011, 50, 3751–3754. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Brennan, J.D.; Li, Y. Characterizing the secondary structure and identifying functionally essential nucleotides of pH6DZ1, a fluorescence-signaling and RNA-cleaving deoxyribozyme. Biochemistry 2005, 44, 12066–12076. [Google Scholar] [CrossRef] [PubMed]
- Tram, K.; Kanda, P.; Li, Y. Lighting up RNA-Cleaving DNAzymes for Biosensing. J. Nucleic Acids 2012, 2012, 958683. [Google Scholar] [CrossRef] [PubMed]
- He, S.N.; Qu, L.; Shen, Z.F.; Tan, Y.; Zeng, M.Y.; Liu, F.; Jiang, Y.Y.; Li, Y.F. Highly Specific Recognition of Breast Tumors by an RNA-Cleaving Fluorogenic DNAzyme Probe. Anal. Chem. 2015, 87, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, S.D.; Ali, M.M.; Salena, B.J.; Li, Y. A sensitive DNA enzyme-based fluorescent assay for bacterial detection. Biomolecules 2013, 3, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, S.D.; Ali, M.M.; Kanda, P.; Li, Y.F. Detection of Bacteria Using Fluorogenic DNAzymes. J. Vis. Exp. 2012, 28. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.F.; Wu, Z.S.; Chang, D.R.; Zhang, W.Q.; Tram, K.; Lee, C.; Kim, P.; Salena, B.J.; Li, Y.F. A Catalytic DNA Activated by a Specific Strain of Bacterial Pathogen. Angew. Chem. Int. Ed. 2016, 55, 2431–2434. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Liu, F.; Zheng, J.; Zeng, M.Y.; Jiang, Y.Y. A Catalytic DNA Probe with Stem-loop Motif for Human T47D Breast Cancer Cells. Anal. Sci. 2015, 31, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Kandadai, S.A.; Li, Y.F. Characterization of pH3DZ1—An RNA-cleaving deoxyribozyme with optimal activity at pH 3. Can. J. Chem. 2007, 85, 261–273. [Google Scholar] [CrossRef]
- Kandadai, S.A.; Li, Y. Characterization of a catalytically efficient acidic RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2005, 33, 7164–7175. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.C.F.; Kwan, S.O.; Li, Y.F. Characterization of non-8–17 sequences uncovers structurally diverse RNA-cleaving deoxyribozymes. Mol. Biosyst. 2011, 7, 2139–2146. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Breaking up is easy to do (if you’re a DNA enzyme that cleaves RNA). Chem. Biol. 2004, 11, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Chiuman, W.; Brennan, J.D.; Li, Y.F. Catalysis and rational engineering of trans-acting pH6DZ1, an RNA-cleaving and fluorescence-signaling deoxyribozyme with a four-way junction structure. ChemBioChem 2006, 7, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Cao, Z.H.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Kandadai, S.A.; Mok, W.W.K.; Ali, M.M.; Li, Y. Characterization of an RNA-Cleaving Deoxyribozyme with Optimal Activity at pH 5. Biochemistry 2009, 48, 7383–7391. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Lu, Y. Improving fluorescent DNAzyme biosensors by combining inter- and intramolecular quenchers. Anal. Chem. 2003, 75, 6666–6672. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Mei, S.H.J.; Brennan, J.D.; Li, Y.F. Assemblage of signaling DNA enzymes with intriguing metal-ion specificities and pH dependences. J. Am. Chem. Soc. 2003, 125, 7539–7545. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA Enzyme with Mg2+-Dependent RNA Phosphoesterase Activity. Chem. Biol. 1995, 2, 655–660. [Google Scholar] [CrossRef]
- Fey, S.; Elling, L.; Kragl, U. The cofactor Mg2+—A key switch for effective continuous enzymatic production of GDP-mannose using recombinant GDP-mannose pyrophosphorylase. Carbohydr. Res. 1997, 305, 475–481. [Google Scholar] [CrossRef]
- Faulhammer, D.; Famulok, M. Characterization and divalent metal-ion dependence of in vitro selected deoxyribozymes which cleave DNA/RNA chimeric oligonucleotides. J. Mol. Biol. 1997, 269, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Billen, L.P.; Li, Y.F. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Bio. 2002, 9, 507–517. [Google Scholar] [CrossRef]
- Li, Y.F.; Breaker, R.R. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Peracchi, A. Preferential activation of the 8–17 deoxyribozyme by Ca2+ ions—Evidence for the identity of 8–17 with the catalytic domain of the MG5 deoxyribozyme. J. Biol. Chem. 2000, 275, 11693–11697. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Yarus, M. Finding specific RNA motifs: Function in a zeptomole world? RNA 2003, 9, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; De Sterck, H.; Markel, R.; Smit, S.; Oshmyansky, A.; Yarus, M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005, 33, 5924–5935. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Unrau, P.J.; Bartel, D.P. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 1997, 4, 767–774. [Google Scholar] [CrossRef]
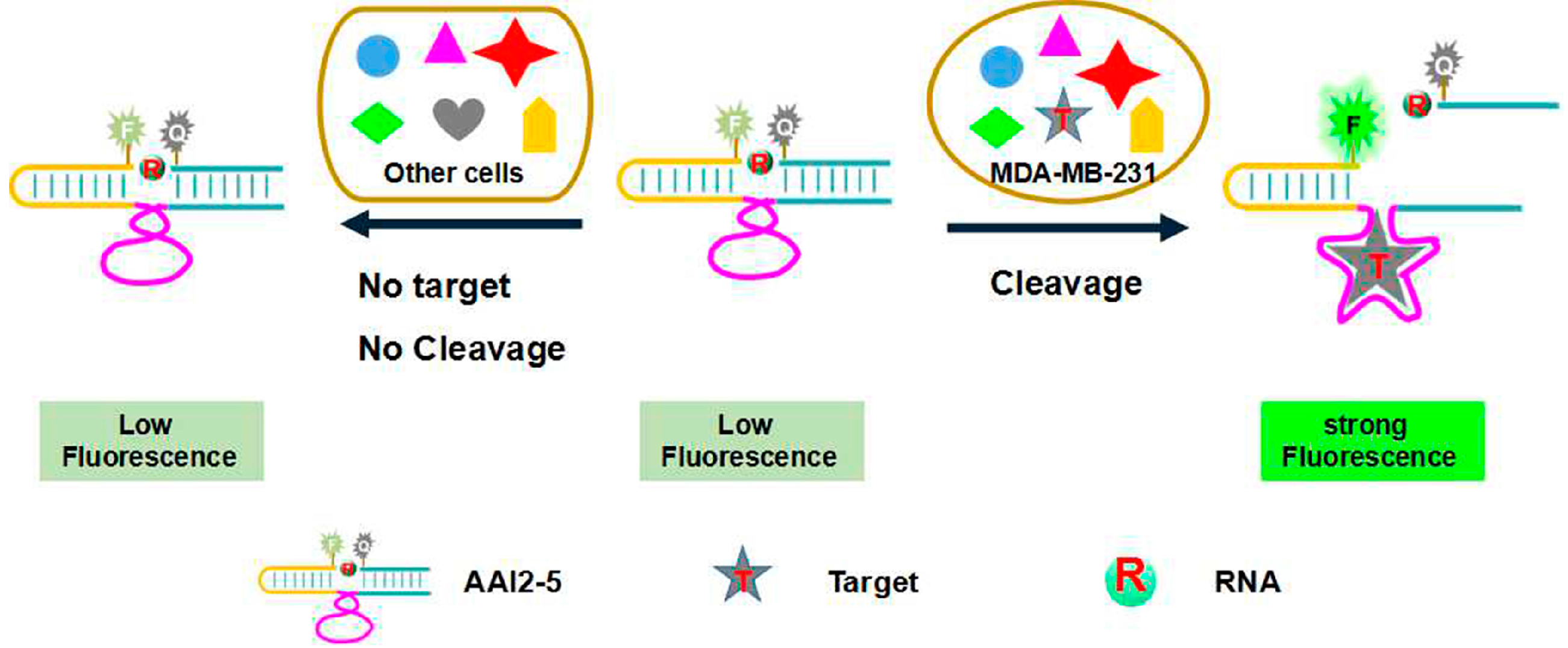





| Name | Sequences 5′-3′ |
|---|---|
| Substrate | 5′-ACTCTTCCTAGC-FRQ-GGTTCGATCAAGA-3′ |
| AL | 5′-CACGGATCCTGACAAGGGAACGGTTAGATCTGATACCTTAGCGAAGGTGTGGTTGGCCAGCTCCGTCCG-3′ |
| Template 1 | 5′-CTAGGAAGAGTCGGACGGAGCTG-3′ |
| Template 2 | 5′-CTAGGAAGAGTGCCAACCACACC-3′ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, P.; He, S.; Mao, Y.; Qu, L.; Liu, F.; Tan, C.; Jiang, Y.; Tan, Y. The Optimization and Characterization of an RNA-Cleaving Fluorogenic DNAzyme Probe for MDA-MB-231 Cell Detection. Sensors 2017, 17, 650. https://doi.org/10.3390/s17030650
Xue P, He S, Mao Y, Qu L, Liu F, Tan C, Jiang Y, Tan Y. The Optimization and Characterization of an RNA-Cleaving Fluorogenic DNAzyme Probe for MDA-MB-231 Cell Detection. Sensors. 2017; 17(3):650. https://doi.org/10.3390/s17030650
Chicago/Turabian StyleXue, Pengcheng, Shengnan He, Yu Mao, Long Qu, Feng Liu, Chunyan Tan, Yuyang Jiang, and Ying Tan. 2017. "The Optimization and Characterization of an RNA-Cleaving Fluorogenic DNAzyme Probe for MDA-MB-231 Cell Detection" Sensors 17, no. 3: 650. https://doi.org/10.3390/s17030650





