Probing the Kinetic Anabolism of Poly-Beta-Hydroxybutyrate in Cupriavidus necator H16 Using Single-Cell Raman Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Culture Medium
2.2. Bacterial Culture and Sampling
2.3. Experimental Setup and Raman Measurements
2.4. Spectral Data Analysis
3. Results and Discussion
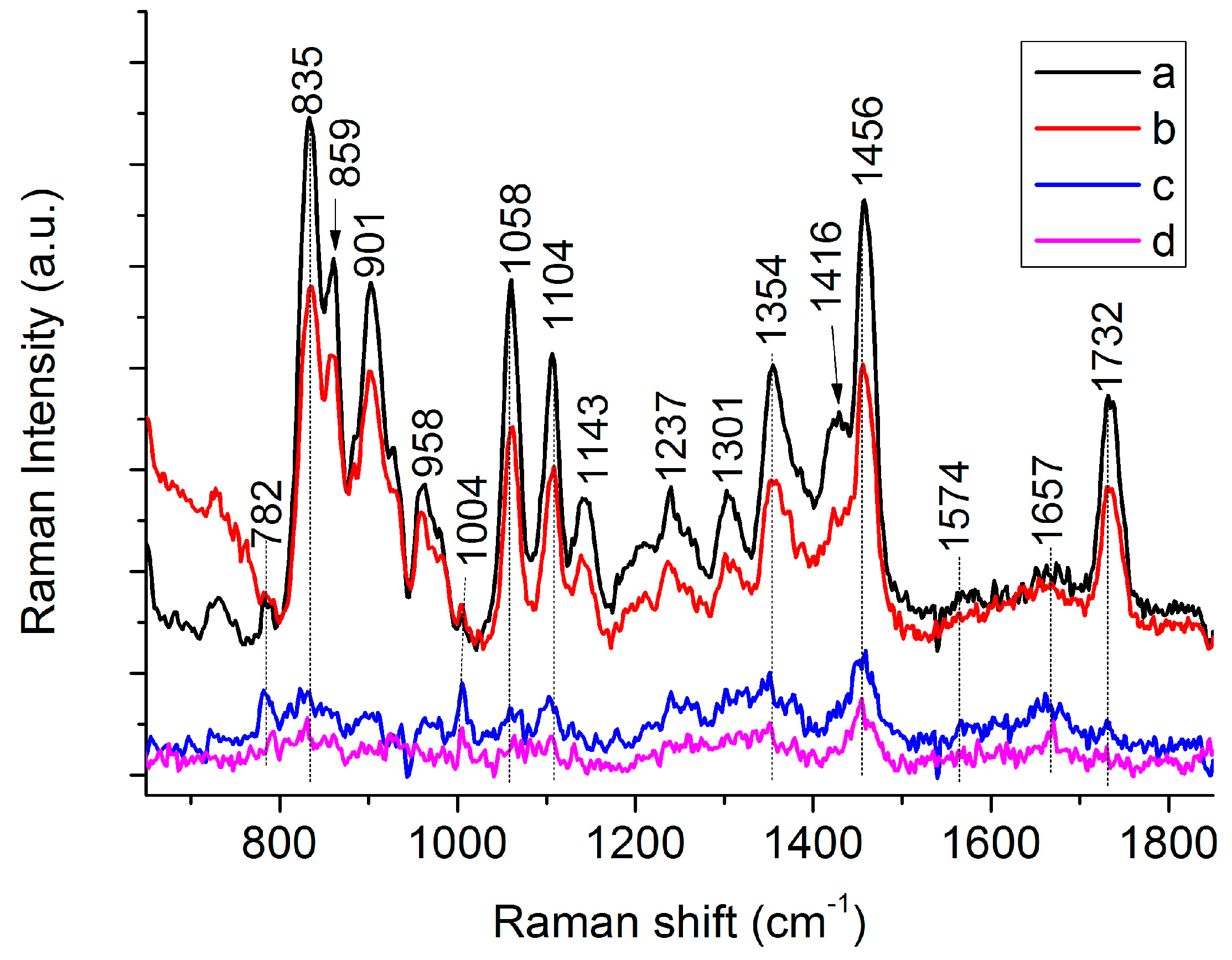
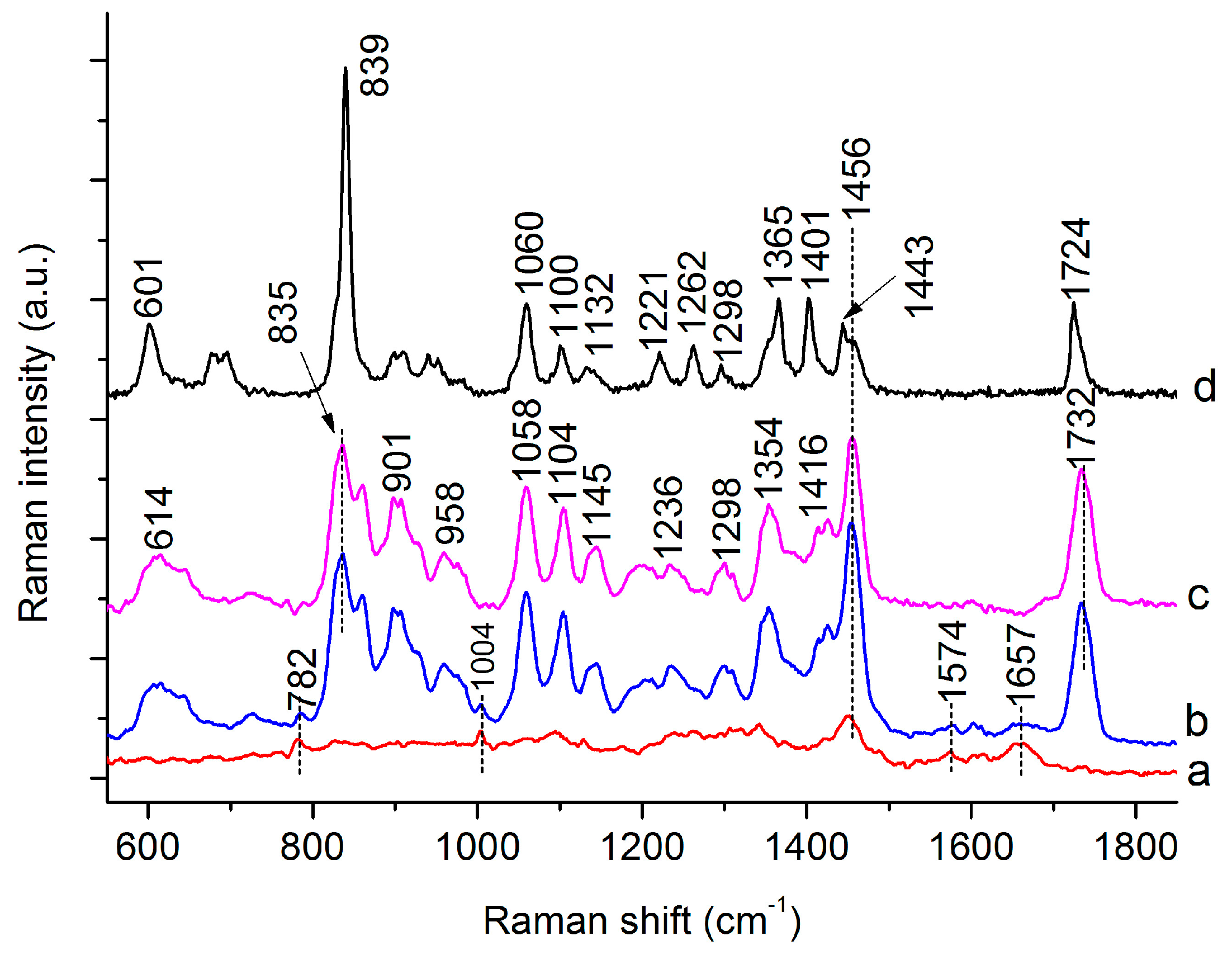
3.1. Raman Spectra of Single C. necator Cells
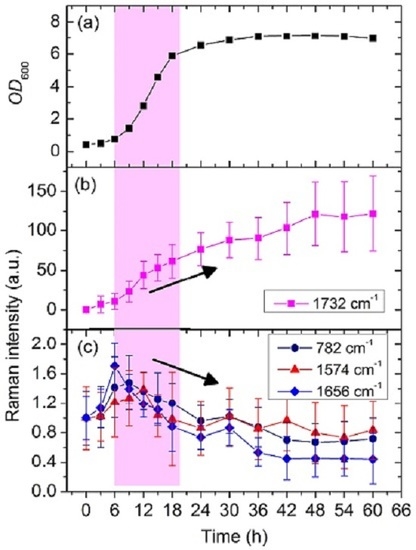
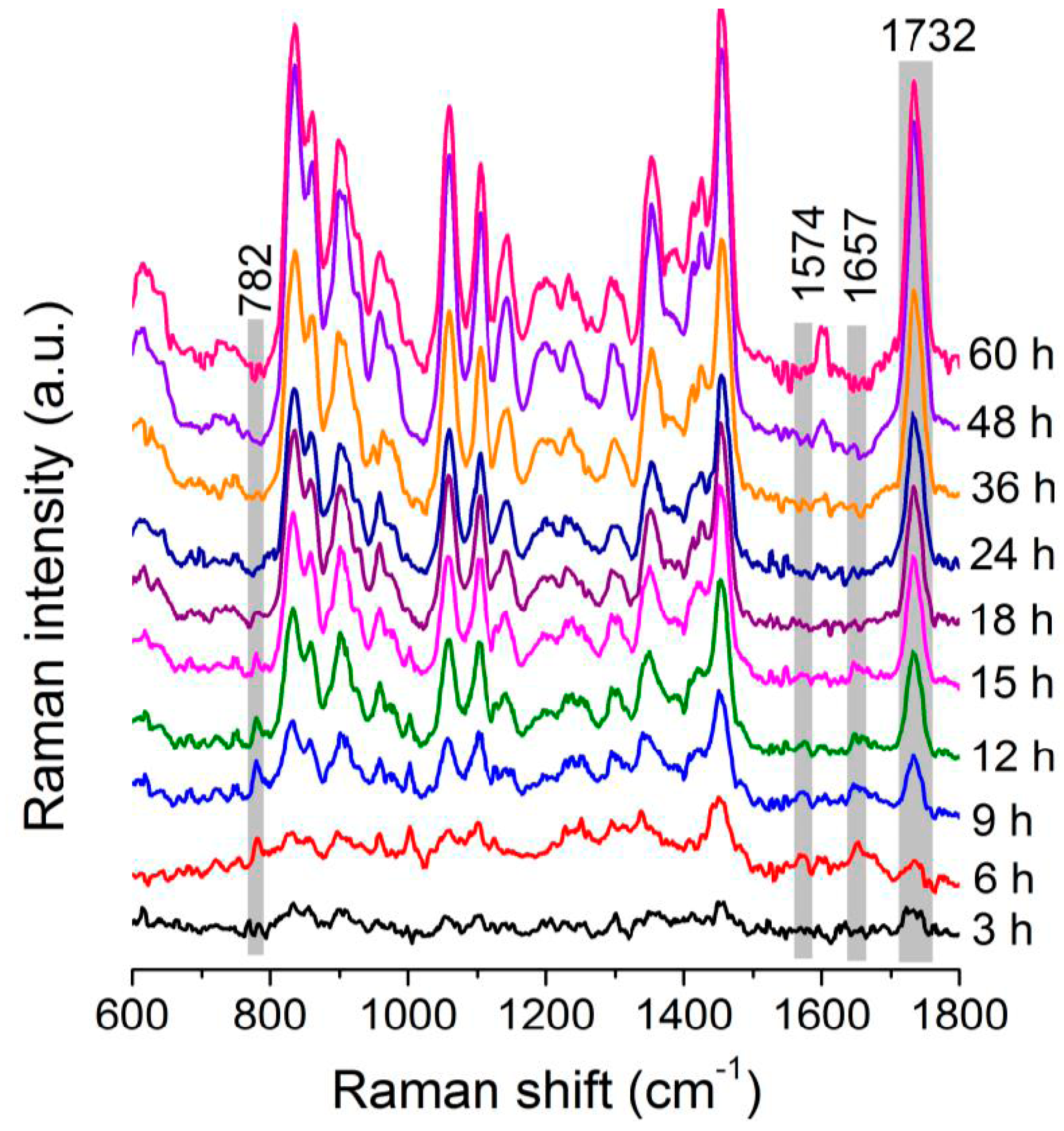
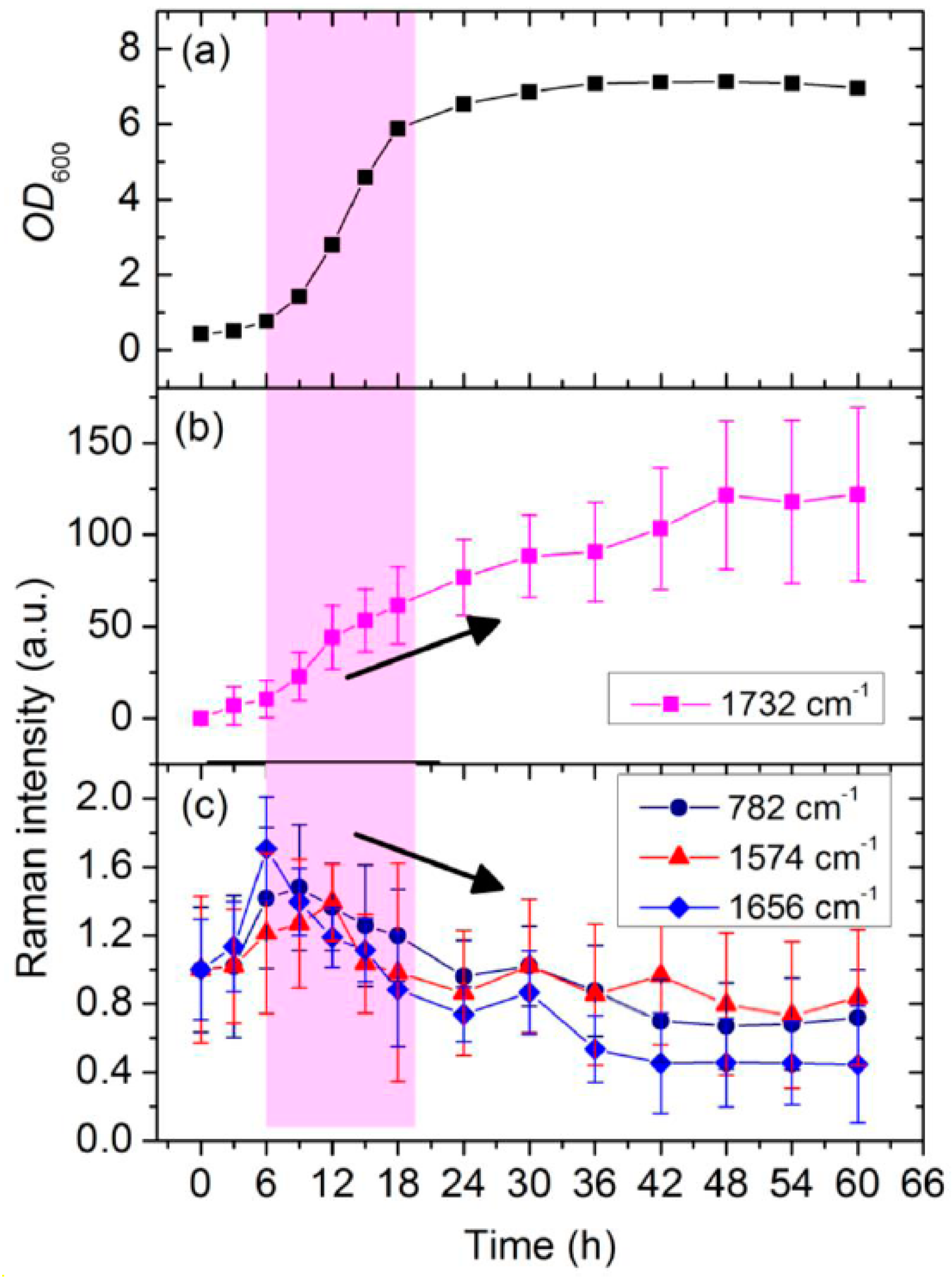
3.2. Dynamic Changes in Biomolecule Level inside C. necator H16 Cells during Batch Culture
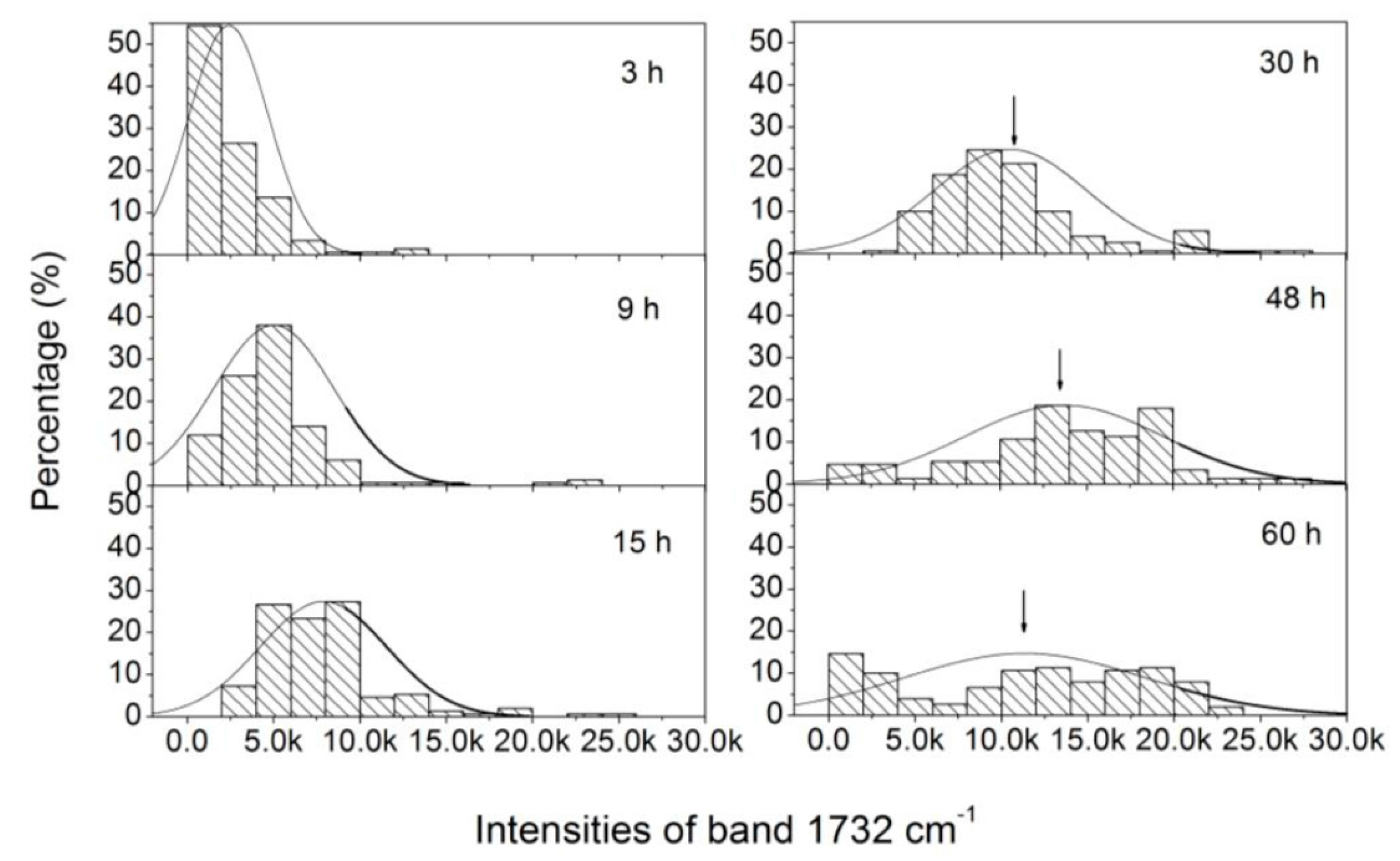
3.3. Analysis of the Heterogeneity of PHB Production at the Single-Cell Level
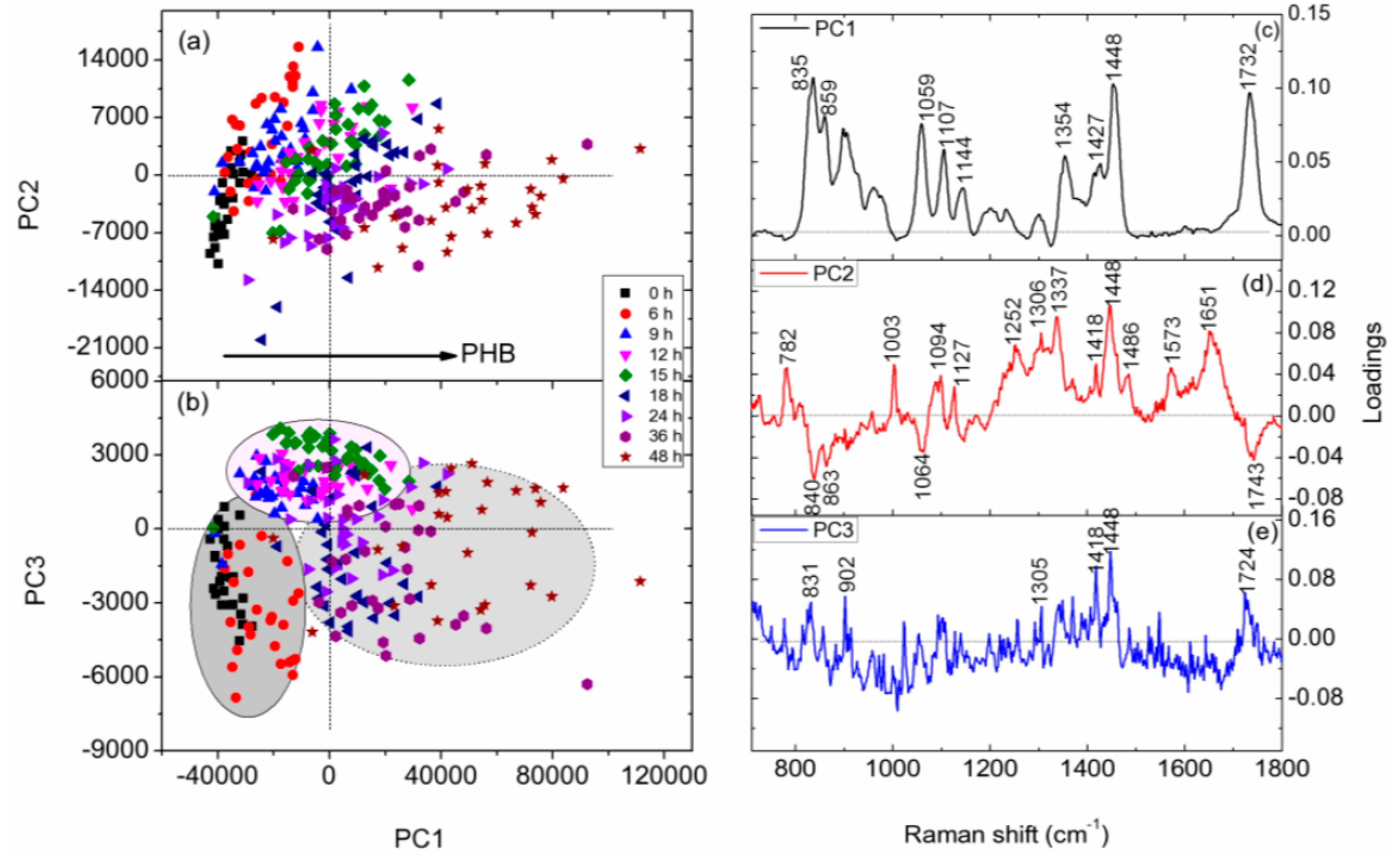
3.4. PCA of Raman Spectra of Individual C. necator Cells at Different Growth Stages
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gao, S.; Feng, J.; Sun, L.H.; Tian, H. The Evolvement of Co-Production of Biodegradable Biological Material PHB. In Proceedings of the 2015 4th International Conference on Sensors, Measurement and Intelligent Materials, Shenzhen, China, 27–28 December 2015; Volume 43, pp. 728–731.
- Rehm, B.H. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 2009, 38, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar] [PubMed]
- Choi, J.; Lee, S.Y. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl. Microbiol. Biotechnol. 1999, 51, 13–21. [Google Scholar] [CrossRef]
- York, G.M.; Lupberger, J.; Tian, J.; Lawrence, A.G.; Stubbe, J.; Sinskey, A.J. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 2003, 185, 3788–3794. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Sinskey, A.J.; Stubbe, J. Kinetic studies of polyhydroxybutyrate granule formation in wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 2005, 187, 3814–3824. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.; Fricke, W.F.; Reinecke, F.; Kusian, B.; Liesegang, H.; Cramm, R.; Eitinger, T.; Ewering, C.; Potter, M.; Schwartz, E.; et al. Genome sequence of the bioplastic-producing “knallgas” bacterium ralstonia eutropha H16. Nat. Biotechnol. 2006, 24, 1257–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y. Deciphering bioplastic production. Nat. Biotechnol. 2006, 24, 1227–1229. [Google Scholar] [CrossRef] [PubMed]
- Cramm, R. Genomic view of energy metabolism in ralstonia eutropha H16. J. Mol. Microbiol. Biotechnol. 2009, 16, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Batcha, A.F.; Prasad, D.M.; Khan, M.R.; Abdullah, H. Biosynthesis of poly(3-hydroxybutyrate) (PHB) by cupriavidus necator H16 from jatropha oil as carbon source. Bioprocess Biosyst. Eng. 2014, 37, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Peoples, O.P.; Sinskey, A.J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 1989, 264, 15298–15303. [Google Scholar] [PubMed]
- Yu, H.M.; Yin, J.; Li, H.Q.; Yang, S.L.; Shen, Z.Y. Construction and selection of the novel recombinant Escherichia coli strain for poly(beta-hydroxybutyrate) production. J. Biosci. Bioeng. 2000, 89, 307–311. [Google Scholar] [CrossRef]
- Yoo, S.; Kim, W.S. Cybernetic model for synthesis of poly-beta-hydroxybutyric acid in alcaligenes-eutrophus. Biotechnol. Bioeng. 1994, 43, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Karr, D.B.; Waters, J.K.; Emerich, D.W. Analysis of poly-beta-hydroxybutyrate in rhizobium-japonicum bacteroids by ion-exclusion high-pressure liquid-chromatography and UV detection. Appl. Environ. Microbiol. 1983, 46, 1339–1344. [Google Scholar] [PubMed]
- Petry, R.; Schmitt, M.; Popp, J. Raman spectroscopy—A prospective tool in the life sciences. ChemPhysChem 2003, 4, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.E.; Griffiths, R.I.; Thompson, I.P.; Bailey, M.J.; Whiteley, A.S. Raman microscopic analysis of single microbial cells. Anal. Chem. 2004, 76, 4452–4458. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wang, G.; Liao, W.; Yao, H.; Huang, S.; Li, Y.Q. Intracellular ethanol accumulation in yeast cells during aerobic fermentation: A raman spectroscopic exploration. Lett. Appl. Microbiol. 2010, 51, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Gelder, J.D.; Willemse-Erix, D.; Scholtes, M.J.; Sanchez, J.I.; Maquelin, K.; Vandenabeele, P.; Boever, P.D.; Puppels, G.J.; Moens, L.; Vos, P.D. Monitoring poly(3-hydroxybutyrate) production in Cupriavidus necator DSM 428 (H16) with raman spectroscopy. Anal. Chem. 2008, 80, 2155–2160. [Google Scholar] [CrossRef] [PubMed]
- Hermelink, A.; Stammler, M.; Naumann, D. Observation of content and heterogeneity of poly-beta-hydroxybutyric acid (PHB) in legionella bozemanii by vibrational spectroscopy. Analyst 2011, 136, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Dinno, M.A.; Li, Y.Q. Near-infrared raman spectroscopy of single optically trapped biological cells. Opt. Lett. 2002, 27, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.W. Recent advances in laser tweezers Raman spectroscopy (LTRS) for label-free analysis of single cells. J. Biophotonics 2013, 6, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Redding, B.; Schwab, M.J.; Pan, Y.L. Raman spectroscopy of optically trapped single biological micro-particles. Sensors 2015, 15, 19021–19046. [Google Scholar] [CrossRef] [PubMed]
- Sheu, D.S.; Chen, W.M.; Lai, Y.W.; Chang, R.C. Mutations derived from the thermophilic polyhydroxyalkanoate synthase PhaC enhance the thermostability and activity of PhaC from cupriavidus necator H16. J. Bacterio. 2012, 194, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- La, R. Encyclopedia of Soil Science; CRC Press: Cleveland, OH, USA, 2005; Volume 2. [Google Scholar]
- Furukawa, T.; Sato, H.; Murakami, R.; Zhang, J.; Noda, I.; Ochiai, S.; Ozaki, Y. Raman microspectroscopy study of structure, dispersibility, and crystallinity of poly(hydroxybutyrate)/poly(l-lactic acid) blends. Polymer 2006, 47, 3132–3140. [Google Scholar] [CrossRef]
- Notingher, I.; Verrier, S.; Haque, S.; Polak, J.M.; Hench, L.L. Spectroscopic study of human lung epithelial cells (A549) in culture: Living cells versus dead cells. Biopolymers 2003, 72, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Izumi, C.M.S.; Temperini, M.L.A. Ft-raman investigation of biodegradable polymers: Poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate). Vib. Spectrosc. 2010, 54, 127–132. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Khanna, S.; Srivastava, A.K. Statistical media optimization studies for growth and PHB production by ralstonia eutropha. Process Biochem. 2005, 40, 2173–2182. [Google Scholar] [CrossRef]
- Masamune, S.; Walsh, C.T.; Sinskey, A.J.; Peoples, O.P. Poly-(R)-3-hydroxybutyrate (PHB) biosynthesis—Mechanistic studies on the biological claisen condensation catalyzed by beta-ketoacyl thiolase. Pure Appl. Chem. 1989, 61, 303–312. [Google Scholar] [CrossRef]
- Klug, W.S.; Cummings, M.R.; Spencer, C.; Palladino, M.A. Concepts of Genetics; Benjamin Cummings: San Francisco, CA, USA, 2011. [Google Scholar]
| Peaks (cm−1) | Assignment a | ||
|---|---|---|---|
| 0 h Cell | 24 h Cell | PHB | |
| 782 | 782 | G, U of RNA [19,27] | |
| 832 | 835 | 839 | C-O-C str. of PHB [26,28], “exposed” Tyr of cells [29] |
| 859 | 859 | C-O-C str. of PHB [28], “buried” Tyr of cells [27,29] | |
| 901 | 899 | υ(COC) [19] | |
| 958 | 952 | C-C str. and CH3 rocking of PHB [26,28] | |
| 1004 | 1004 | Phe [27,29] | |
| 1058 | 1059 | C-O str. [26,28] | |
| 1094 | PO2- str. of DNA [27,29] | ||
| 1104 | 1100 | C-O-C sym. str. of PHB [26,28] | |
| 1129 | 1143 | 1132 | 1129, C-N str. of proteins [27,29] |
| 1143, (C-N), (C-C) str. of proteins [27,29] | |||
| 1132, C-O-C sym. str. of PHB [26,28] | |||
| 1237 | 1221 | 1237, amide III [19,27] | |
| 1221, C-O-C asymmetric str. [26,28] | |||
| 1262 | 1262 | 1262 | C-O-C str. and CH def. [26,28] |
| Helical conformation (C) of PHB [26,28] | |||
| 1305 | 1301 | 1294 | CH2 twist of lipids, CH def. of PHB [26,28] |
| 1342 | A, G of nucleic acid and C–H of proteins. [19,27] | ||
| 1354 | 1365 | Sym. str. CH3 and CH def of PHB [26,28] | |
| 1416 | 1401 | CH3 sym. def. [26,28] | |
| 1453 | 1456 | 1443 | CH2/CH3 [26,28] |
| 1574 | 1574 | G, A of DNA [19,27] | |
| 1657 | 1657 | Amide I [19,27] | |
| 1732 | 1724 | 1732 C=O str. of PHB (Amorphous) [26]/C=O str. of lipids [29] | |
| 1724 C=O str. of PHB (Crystalline) [26] | |||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Z.; Peng, L.; Zhang, P.; Li, Y.-Q.; Wang, G. Probing the Kinetic Anabolism of Poly-Beta-Hydroxybutyrate in Cupriavidus necator H16 Using Single-Cell Raman Spectroscopy. Sensors 2016, 16, 1257. https://doi.org/10.3390/s16081257
Tao Z, Peng L, Zhang P, Li Y-Q, Wang G. Probing the Kinetic Anabolism of Poly-Beta-Hydroxybutyrate in Cupriavidus necator H16 Using Single-Cell Raman Spectroscopy. Sensors. 2016; 16(8):1257. https://doi.org/10.3390/s16081257
Chicago/Turabian StyleTao, Zhanhua, Lixin Peng, Pengfei Zhang, Yong-Qing Li, and Guiwen Wang. 2016. "Probing the Kinetic Anabolism of Poly-Beta-Hydroxybutyrate in Cupriavidus necator H16 Using Single-Cell Raman Spectroscopy" Sensors 16, no. 8: 1257. https://doi.org/10.3390/s16081257