Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Camera and Software
2.3. Hyperspectral Images
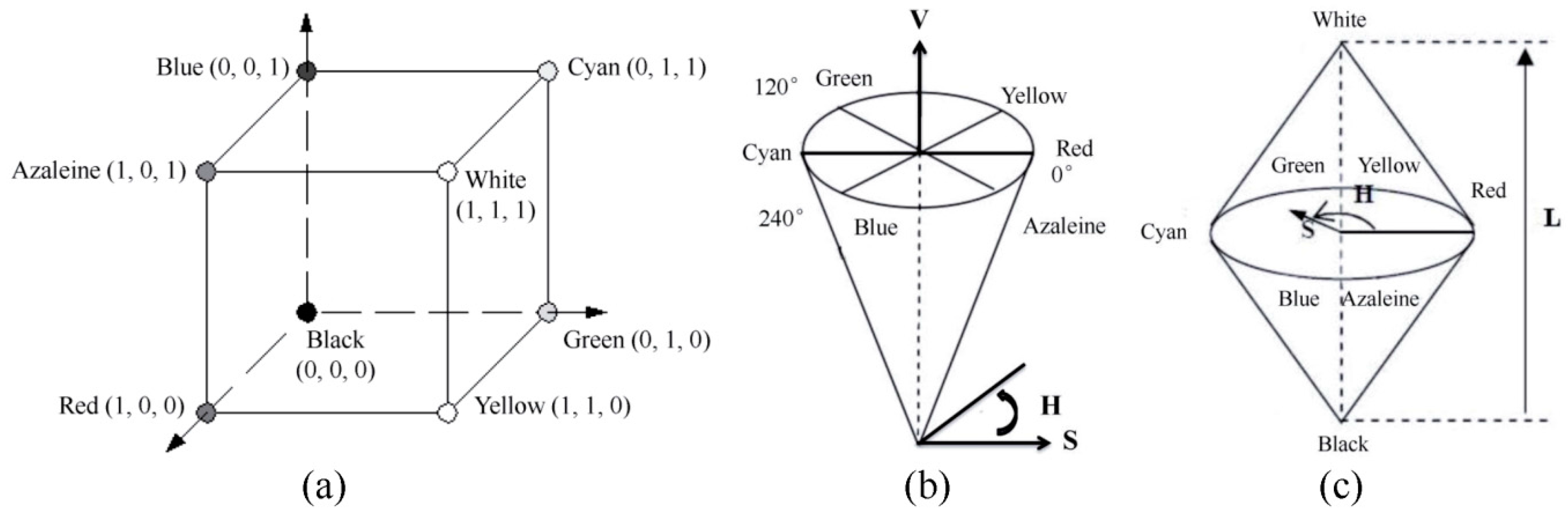
2.4. Conversion from RGB into HSV and HLS Images
2.5. Texture Features Based on GLCM
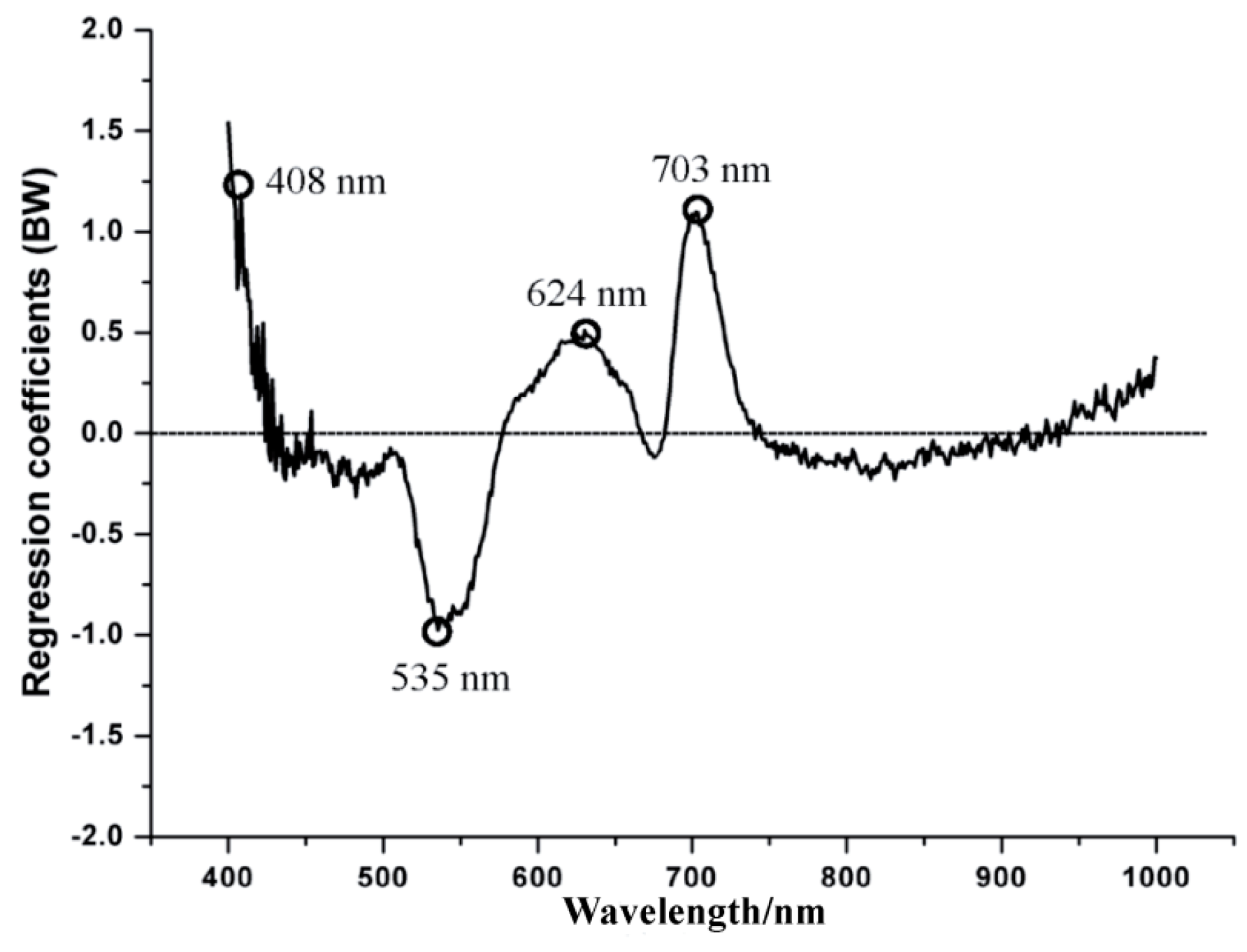
2.6. Regression Coefficients
2.7. Classifiers and Evaluation Performance
2.7.1. Principal Component Analysis
2.7.2. K-Nearest Neighbor and AdaBoost
2.7.3. Model Evaluation
2.8. Study Flow
3. Results and Discussion
3.1. Results Based on Spectrum
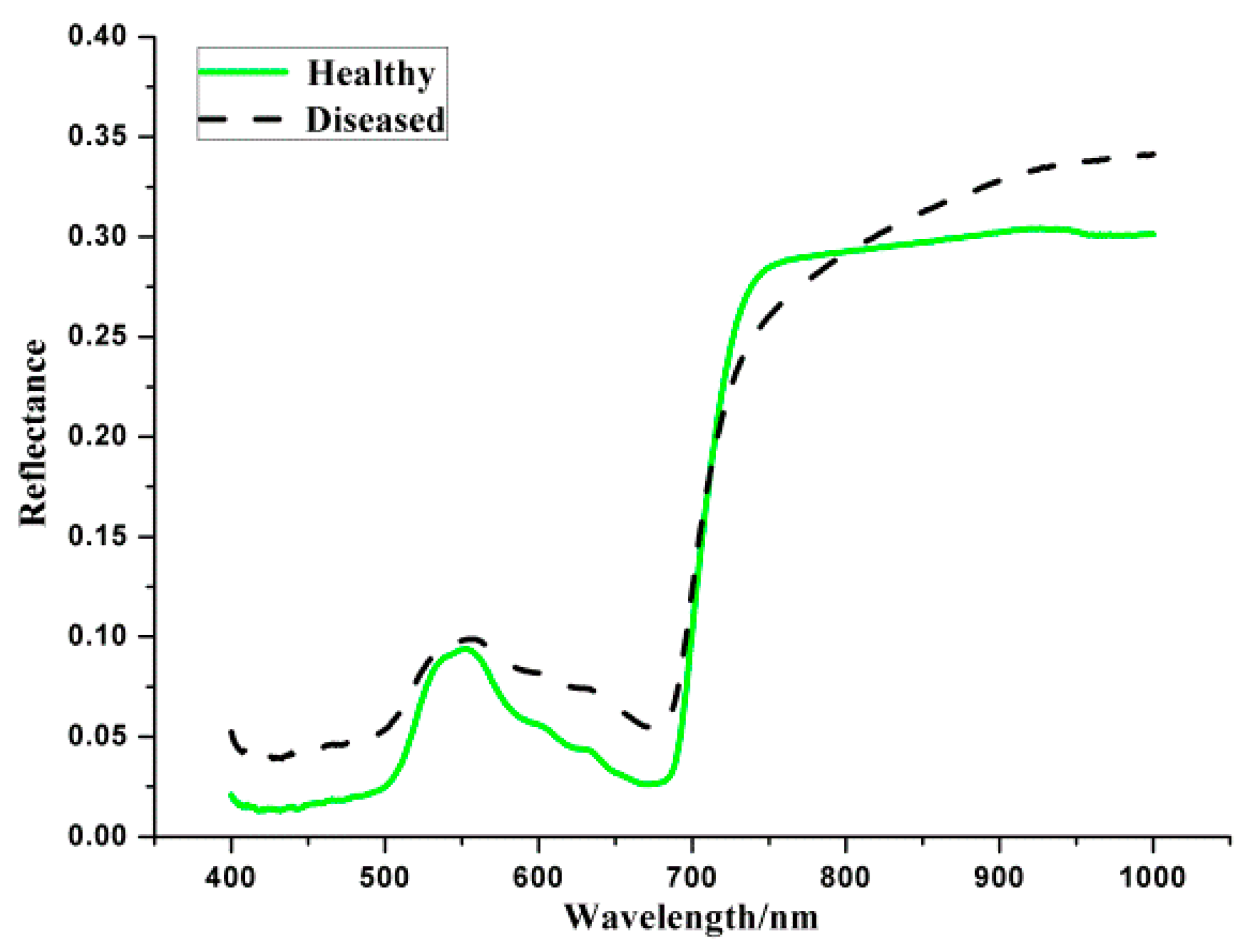
3.1.1. Spectral Reflectance
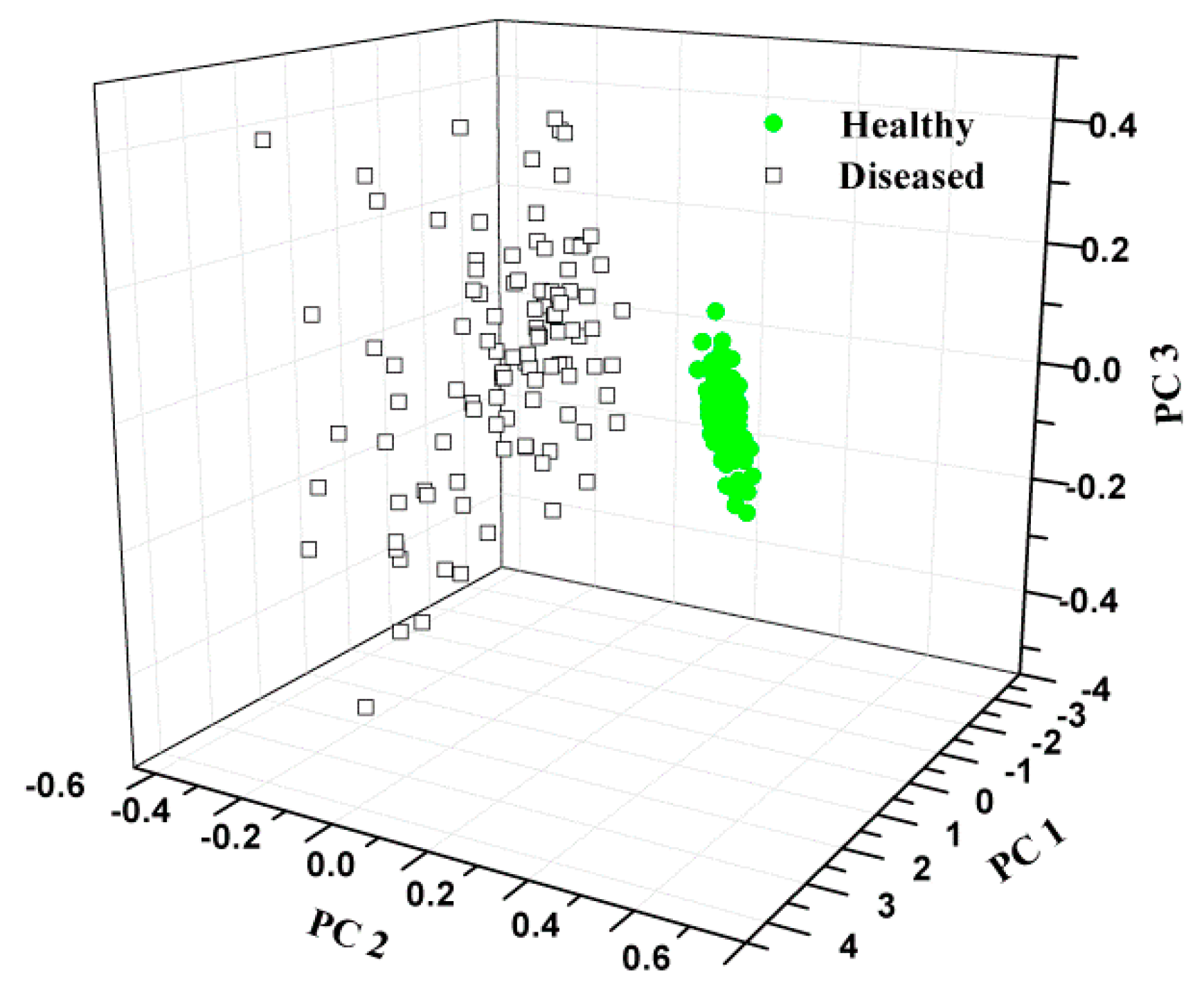
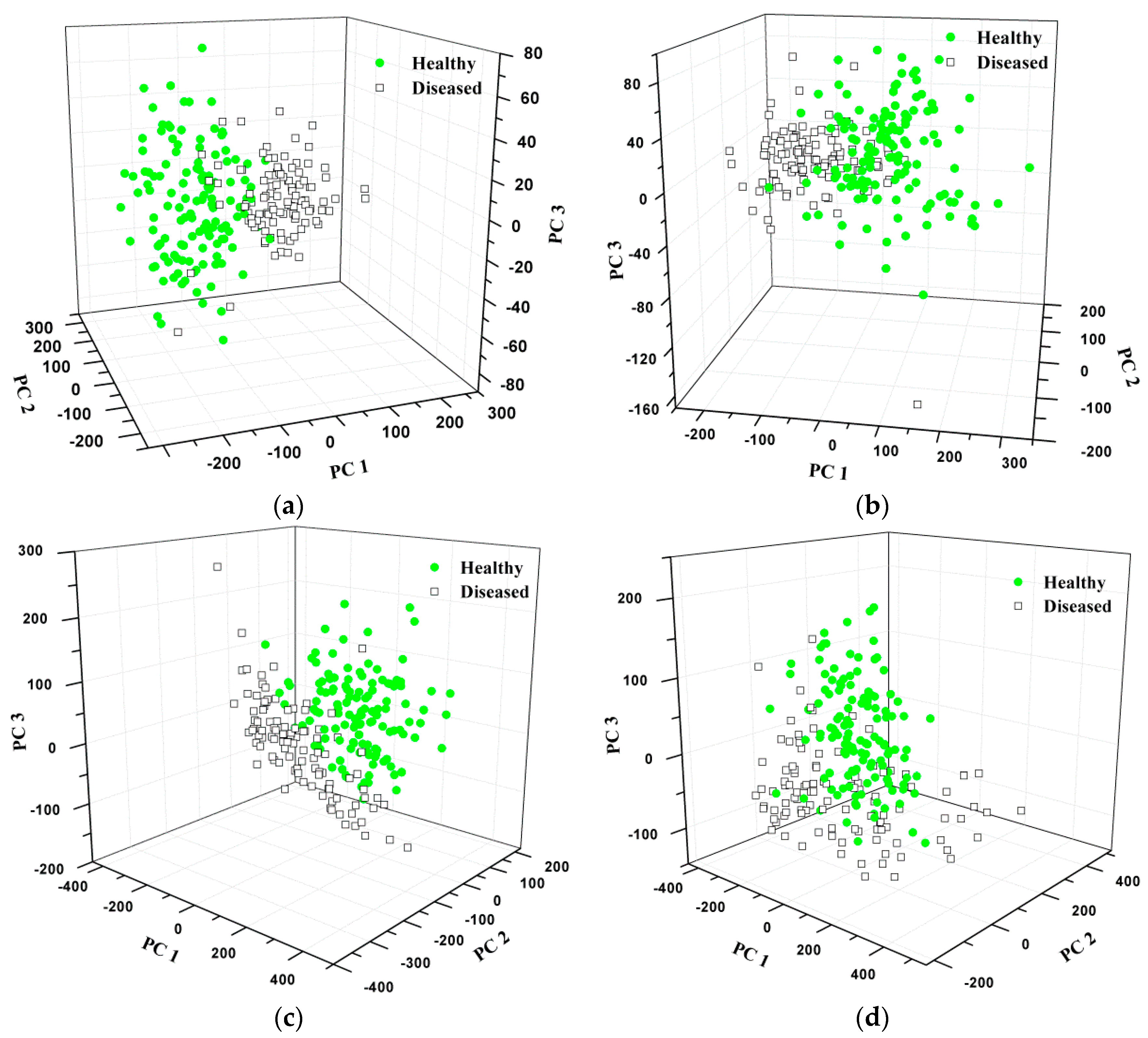
3.1.2. Distribution Based on PCA
3.1.3. Classification Results
3.2. Gray Images
3.3. RGB, HSV and HLS Images
3.4. Results Based on Gray Images
3.4.1. Texture Images
3.4.2. Distribution Based on PCA
3.4.3. Classification Results
3.5. Results Based on RGB Images
3.5.1. Texture Images
3.5.2. Distribution Based on PCA
3.5.3. Classification Results
3.6. Results Based on HSV Images
3.6.1. Texture Images
3.6.2. Distribution Based on PCA
3.6.3. Classification Results
3.7. Results Based on HLS Images
3.7.1. Texture Images
3.7.2. Distribution Based on PCA
3.7.3. Classification Results
3.8. Comparison of Different Images
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xie, C.Q.; Wang, H.L.; Shao, Y.N.; He, Y. Different algorithms for detection of malondialdehyde content in eggplant leaves stressed by grey mold based on hyperspectral imaging techniques. Intell. Autom. Soft Comput. 2015, 21, 395–407. [Google Scholar] [CrossRef]
- Xie, C.Q.; Li, X.L.; Shao, Y.N.; He, Y. Color measurement of tea leaves at different drying periods using hyperspectral imaging technique. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Sun, D.W.; Allen, P. Predicting of moisture, color and pH in cooked, pre-sliced turkey hams by NIR hyperspectral imaging system. J. Food Eng. 2013, 117, 42–51. [Google Scholar] [CrossRef]
- Leiva-Valenzuela, G.A.; Lu, R.F.; Aguilera, J.M. Prediction of firmness and soluble solids content of blueberries using hyperspectral reflectance imaging. J. Food Eng. 2013, 115, 91–98. [Google Scholar] [CrossRef]
- Xie, C.Q.; Li, X.L.; Nie, P.C.; He, Y. Application of time series hyperspectral imaging (TS-HSI) for determining water content within tea leaves during drying. Trans. ASABE 2013, 56, 1431–1440. [Google Scholar]
- Richard, M.; Sven, S.; Sildomar, T.M. Consistency of measurements of wavelength position from hyperspectral imagery: Use of the ferric iron crystal field absorption at similar to 900 nm as an indicator of mineralogy. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2843–2857. [Google Scholar]
- Michael, D.; Geert, V.; Clemen, A.; Michael, W.; Michal, R. New ways to extract archaeological information from hyperspectral pixels. J. Archaeol. Sci. 2014, 52, 84–96. [Google Scholar]
- Huang, W.J.; Lamb, D.W.; Niu, Z.; Zhang, Y.J.; Liu, L.Y.; Wang, J.H. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Steiner, U.; Hillnhütter, C.; Dehne, H.W.; Oerke, E.C. Hyperspectral imaging for small-scale analysis of symptoms caused by different sugar beet diseases. Plant Methods 2013, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bulanon, D.M.; Burks, T.F.; Kim, D.G.; Titenour, M.A. Citrus black spot detection using hypersepctral image analysis. Agric. Eng. Int. 2013, 15, 171–180. [Google Scholar]
- Zhu, F.L.; Zhang, D.R.; He, Y.; Liu, F.; Sun, D.W. Application of visible and near infrared hyperspectral imaging to differentiate between fresh and frozen-thawed fish fillets. Food Bioprocess Technol. 2013, 6, 2931–2937. [Google Scholar] [CrossRef]
- Zheng, C.X.; Sun, D.W.; Zheng, L.Y. Recent applications of image texture for evaluation of food qualities—A review. Trends Food Sci. Technol. 2006, 17, 113–128. [Google Scholar] [CrossRef]
- Zhang, X.L.; Liu, F.; He, Y.; Li, X.L. Application of hyperspectral imaging and chemometric calibrations for variety discrimination of maize seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; ElMasry, G.; Sun, D.W.; Allen, P. Prediction of some quality attributes of lamb meat using near-infrared hyperspectral imaging and multivariate analysis. Anal. Chim. Acta 2012, 714, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.Q.; Wang, J.Y.; Feng, L.; Liu, F.; Wu, D.; He, Y. Study on the early detection of early blight on tomato leaves using hyperspectral imaging technique based on spectroscopy and texture. Spectrosc. Spect. Anal. 2013, 33, 1603–1607. [Google Scholar]
- Wei, X.; Liu, F.; Qiu, Z.J.; Shao, Y.N.; He, Y. Ripeness classification of astringent persimmon using hyperspectral imaging technique. Food Bioprocess Technol. 2014, 7, 1371–1380. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Xie, C.Q.; Shao, Y.N.; Li, X.L.; He, Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; He, Y. Application of successive projections algorithm for variable selection to determine organic acids of plum vinegar. Food Chem. 2009, 115, 1430–1436. [Google Scholar] [CrossRef]
- ElMasry, G.; Sun, D.W.; Allen, P. Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. J. Food Eng. 2012, 110, 127–140. [Google Scholar] [CrossRef]
- Qin, J.W.; Burks, T.F.; Kim, M.S.; Chao, K.L.; Ritenour, M.A. Citrus canker detection using hyperspectral reflectance imaging and PCA-based image classification method. Sensonry Instrum. Food Qual. 2008, 2, 168–177. [Google Scholar] [CrossRef]
- Wu, D.; Feng, L.; Zhang, C.Q.; He, Y. Early detection of botrytis cinerea on eggplant leaves based on visible and near-infrared spectroscopy. Trans. ASABE 2008, 51, 1133–1139. [Google Scholar] [CrossRef]
- Ruiz, J.R.R.; Parello, T.C.; Gomez, R.C. Comparative study of multivariate methods to identify paper finishes using infrared spectroscopy. IEEE Trans. Instrum. Meas. 2012, 61, 1029–1036. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.M.; Tang, Y.Y.; Yan, Z.; Hang, K.Y. ISABoost: A weak classifier inner structure adjusting based AdaBoost algorithm-ISABoost based application in scene categorization. Neurocomputing 2013, 103, 104–113. [Google Scholar] [CrossRef]
- Cheng, W.C.; Jhan, D.M. A self-constructing cascade classifier with AdaBoost and SVM for pedestrian detection. Eng. Appl. Artif. Intell. 2013, 26, 1016–1028. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of online learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Paul, V.; Jones, M. Rapid object detection using a boosted cascade of simple features. Comput. Vis. Pattern Recognit. 2001, 1, 511–518. [Google Scholar]
- Tan, C.; Li, M.L.; Qin, X. Study of the deasibility of distinguishing cigarettes of different brands using an AdaBoost algorithm and near-infrared spectroscopy. Anal. Bioanal. Chem. 2007, 389, 667–674. [Google Scholar] [CrossRef] [PubMed]
- ElMasry, G.; Wang, N.; ElSayed, A.; Ngadi, M. Hyperspectral imaging for nondestructive determination of some quality attributes for strawberry. J. Food Eng. 2007, 81, 98–107. [Google Scholar] [CrossRef]



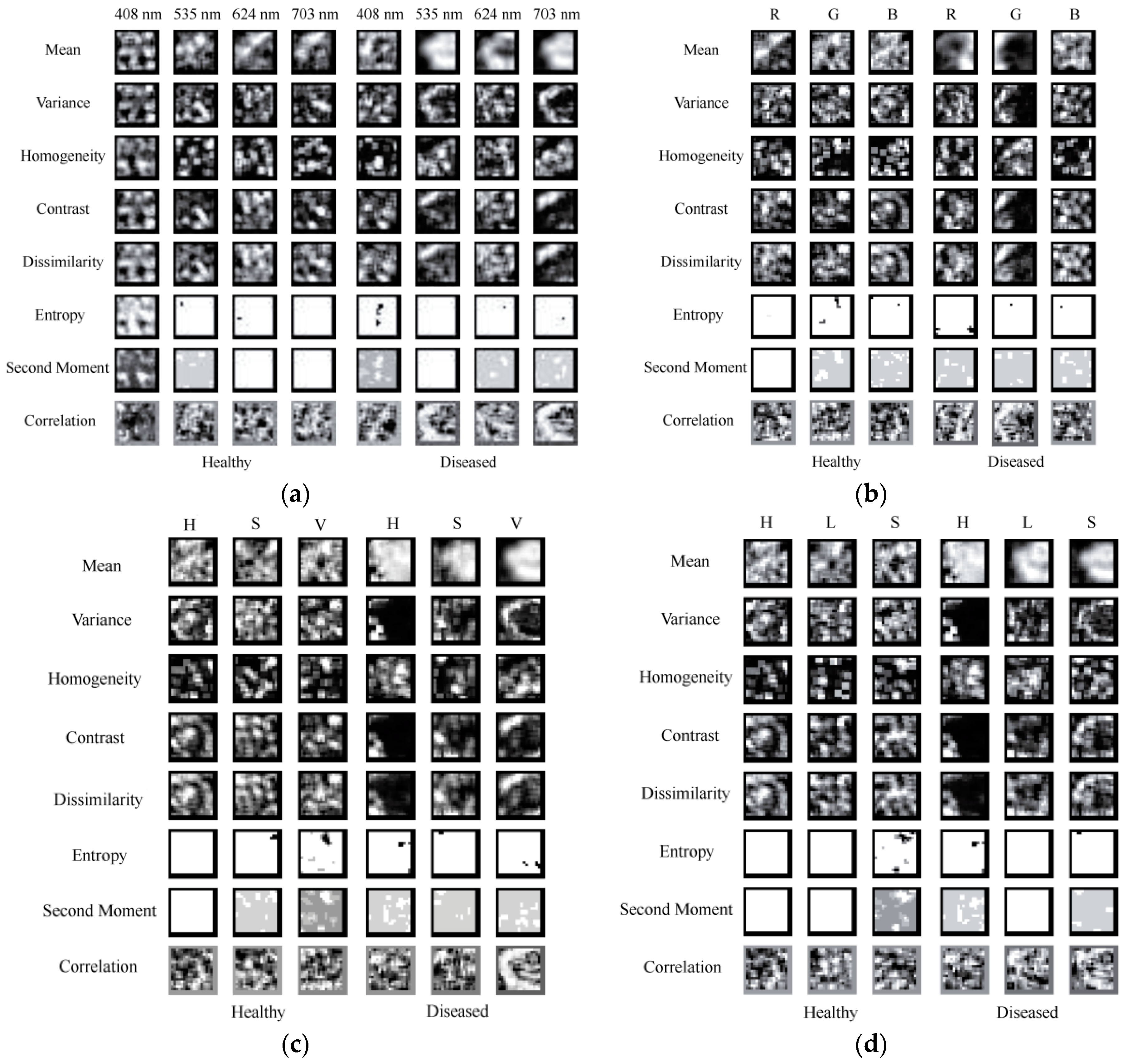
| Textures | Description |
|---|---|
| Mean | The Mean is the average grayscale of all pixels for the image. |
| Variance | The Variance stands for the change of grayscale. |
| Homogeneity | The Homogeneity can measure the uniformity of local grayscale for one image. More uniform of the local grayscale means high value of Homogeneity. |
| Contrast | The Contrast stands for the clarity of texture. The higher the contrast value, the clearer the image. |
| Dissimilarity | The Dissimilarity mainly represents for the difference of grayscale. |
| Entropy | The Entropy is the measurement of the information for one image. No texture feature means Entropy value is 0. |
| Second Moment | The Second Moment reflects the uniformity degree of grayscale. |
| Correlation | The Correlation can measure the level of similarity in the row or column direction. |
| Models | Variables | Training | Testing | ||||
|---|---|---|---|---|---|---|---|
| No. 1 | Correct | CR/% 2 | No. 1 | Correct | CR/% 2 | ||
| KNN | 477 | 157 | 157 | 100 | 78 | 78 | 100 |
| AdaBoost | 477 | 157 | 157 | 100 | 78 | 77 | 98.72 |
| Models | Type | Variables | Training | Testing | ||||
|---|---|---|---|---|---|---|---|---|
| No. 1 | Correct | CR/% 2 | No. 1 | Correct | CR/% 2 | |||
| KNN | Healthy | 32 | 87 | 87 | 100 | 43 | 42 | 97.67 |
| Diseased | 70 | 70 | 100 | 35 | 32 | 91.43 | ||
| All | 157 | 157 | 100 | 78 | 74 | 94.87 | ||
| AdaBoost | Healthy | 32 | 87 | 87 | 100 | 43 | 43 | 100 |
| Diseased | 70 | 70 | 100 | 35 | 34 | 97.14 | ||
| All | 157 | 157 | 100 | 78 | 77 | 98.72 | ||
| Models | Type | Variables | Training | Testing | ||||
|---|---|---|---|---|---|---|---|---|
| No. 1 | Correct | CR/% 2 | No. 1 | Correct | CR/% 2 | |||
| KNN | Healthy | 24 | 87 | 87 | 100 | 43 | 40 | 93.02 |
| Diseased | 70 | 70 | 100 | 35 | 32 | 91.43 | ||
| All | 157 | 157 | 100 | 78 | 72 | 92.31 | ||
| AdaBoost | Healthy | 24 | 87 | 87 | 100 | 43 | 42 | 97.67 |
| Diseased | 70 | 70 | 100 | 35 | 35 | 100 | ||
| All | 157 | 157 | 100 | 78 | 77 | 98.72 | ||
| 10 | Type | Variables | Training | Testing | ||||
|---|---|---|---|---|---|---|---|---|
| No. 1 | Correct | CR/% 2 | No. 1 | Correct | CR/% 2 | |||
| KNN | Healthy | 24 | 87 | 87 | 100 | 43 | 39 | 90.70 |
| Diseased | 70 | 70 | 100 | 35 | 34 | 97.14 | ||
| All | 157 | 157 | 100 | 78 | 73 | 93.59 | ||
| AdaBoost | Healthy | 24 | 87 | 87 | 100 | 43 | 43 | 100 |
| Diseased | 70 | 70 | 100 | 35 | 35 | 100 | ||
| All | 157 | 157 | 100 | 78 | 78 | 100 | ||
| Models | Type | Variables | Training | Testing | ||||
|---|---|---|---|---|---|---|---|---|
| No. 1 | Correct | CR/% 2 | No. 1 | Correct | CR/% 2 | |||
| KNN | Healthy | 24 | 87 | 87 | 100 | 43 | 40 | 93.02 |
| Diseased | 70 | 70 | 100 | 35 | 29 | 82.86 | ||
| All | 157 | 157 | 100 | 78 | 69 | 88.46 | ||
| AdaBoost | Healthy | 24 | 87 | 87 | 100 | 43 | 43 | 100 |
| Diseased | 70 | 70 | 100 | 35 | 33 | 94.29 | ||
| All | 157 | 157 | 100 | 78 | 76 | 97.44 | ||
| Clsssifier | Type | Healthy/% (KNN/AdaBoost) | Diseased/% (KNN/AdaBoost) | All/% (KNN/AdaBoost) | Type | Clsssifier |
|---|---|---|---|---|---|---|
| KNN | Gray image | 97.67/100 | 91.43/97.14 | 94.87/98.72 | Gray image | AdaBoost |
| RGB image | 93.02/97.67 | 91.43/100 | 92.31/98.72 | RGB image | ||
| HSV image | 90.70/100 | 97.14/100 | 93.59/100 | HSV image | ||
| HLS image | 93.02/100 | 82.86/94.29 | 88.46/97.44 | HLS image |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, C.; He, Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors 2016, 16, 676. https://doi.org/10.3390/s16050676
Xie C, He Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors. 2016; 16(5):676. https://doi.org/10.3390/s16050676
Chicago/Turabian StyleXie, Chuanqi, and Yong He. 2016. "Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves" Sensors 16, no. 5: 676. https://doi.org/10.3390/s16050676







