Crop Phenology Detection Using High Spatio-Temporal Resolution Data Fused from SPOT5 and MODIS Products
Abstract
:1. Introduction
2. Study Area and Data
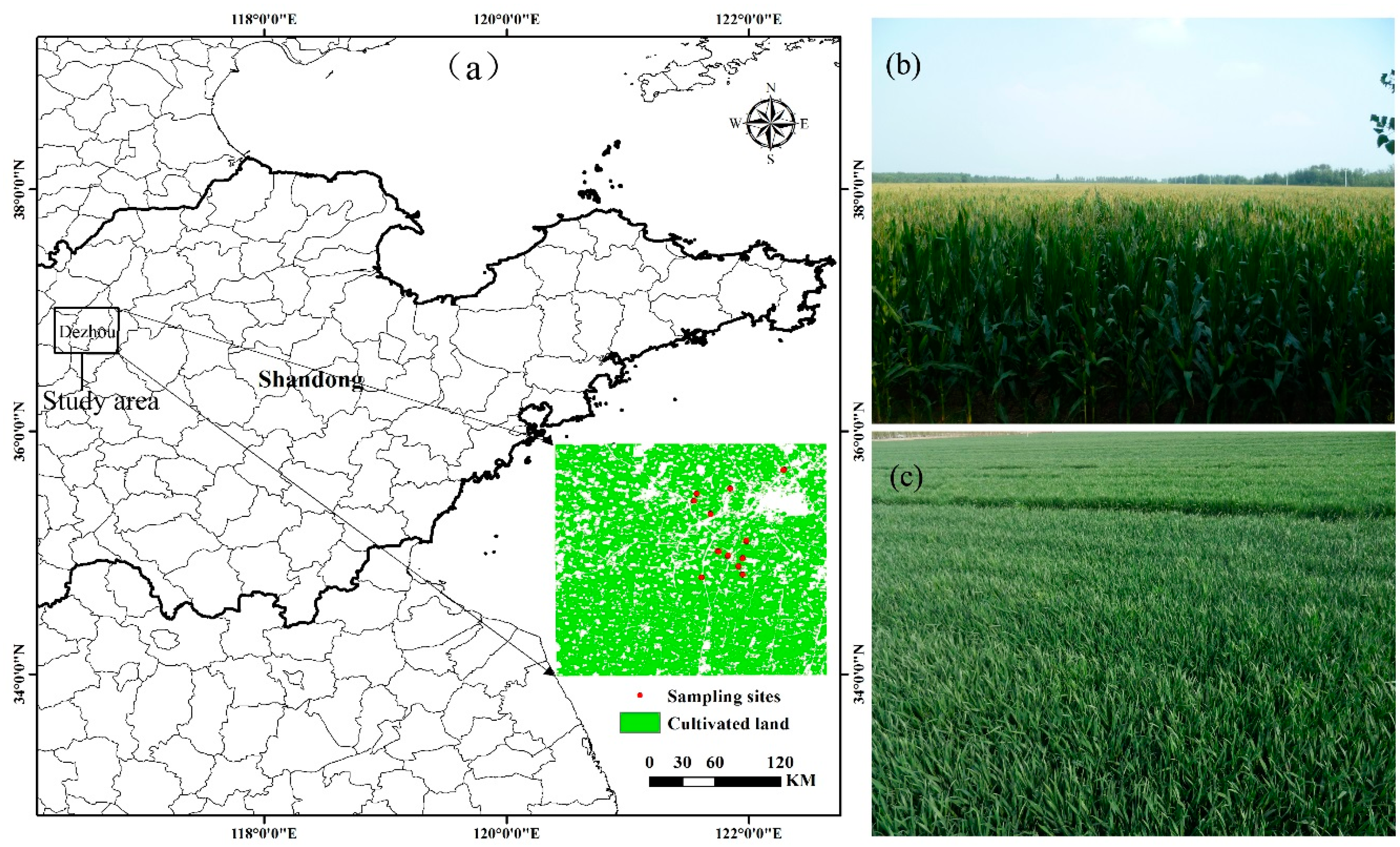
2.1. Study Area
2.2. Data Acquisition and Processing
2.2.1. Ground Data
2.2.2. Satellite Data
2.2.3. Auxiliary Data
3. Methods
3.1. Data Fusion
3.2. Data Smoothing
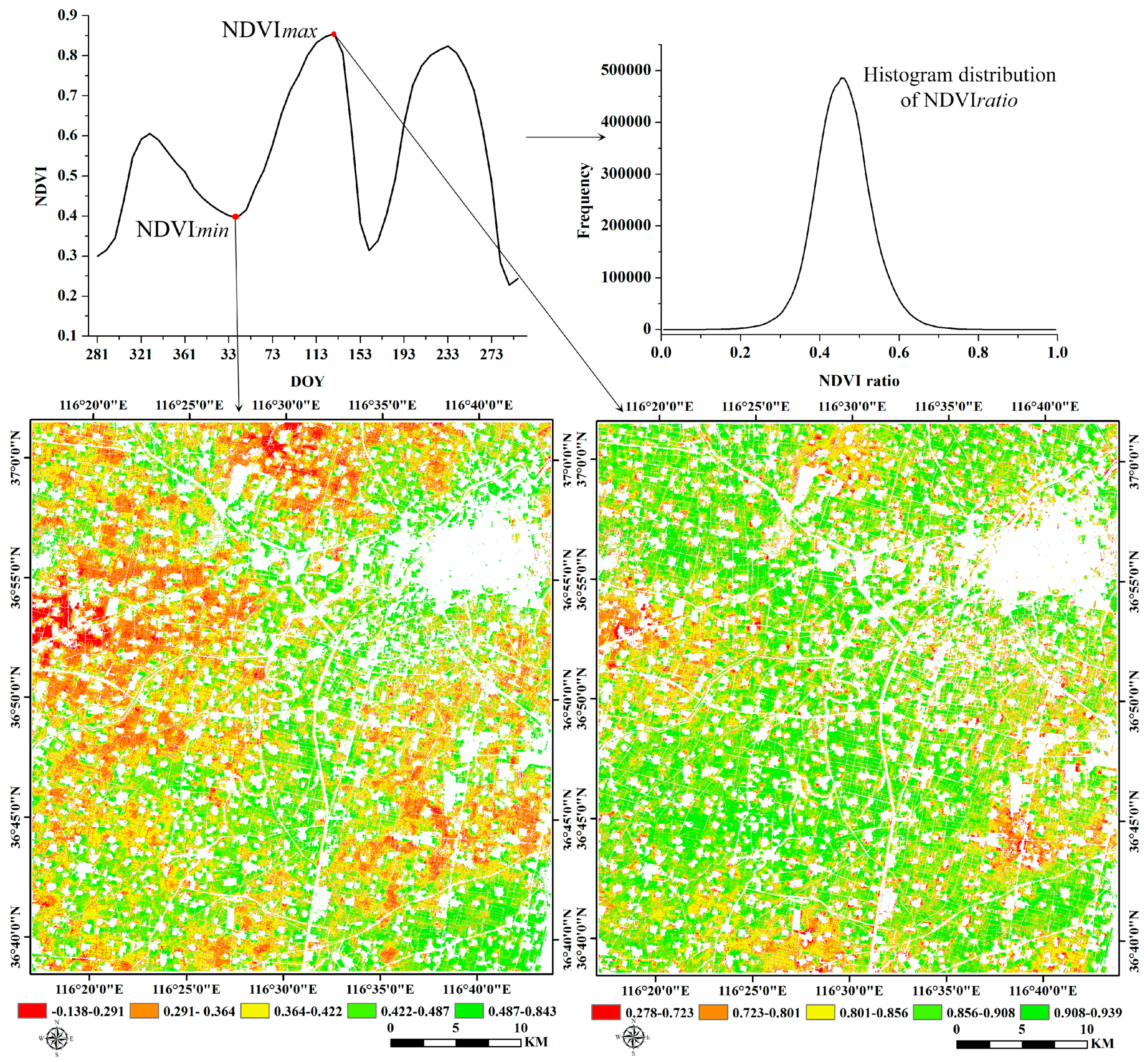
3.3. Phenology Detection
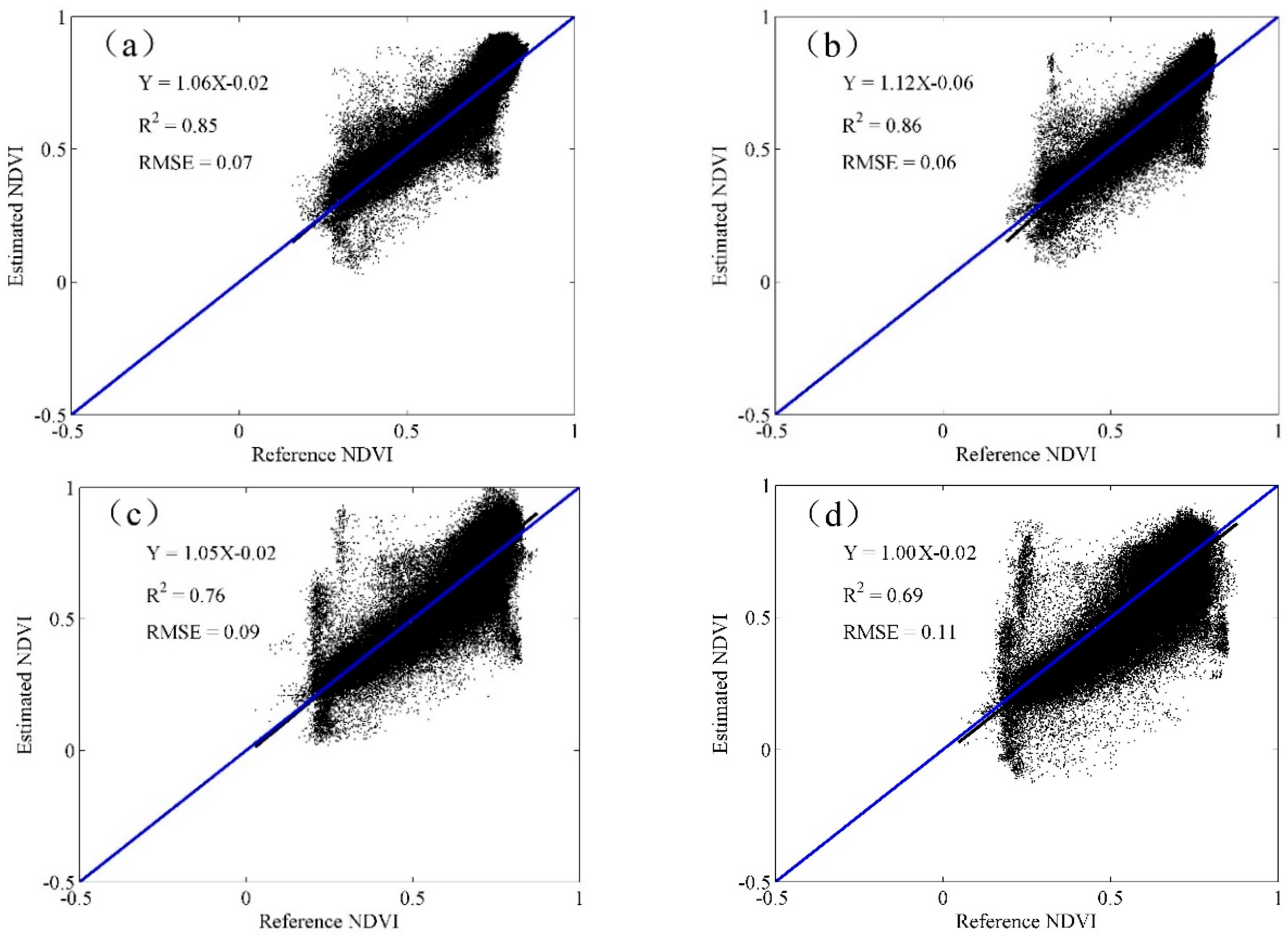
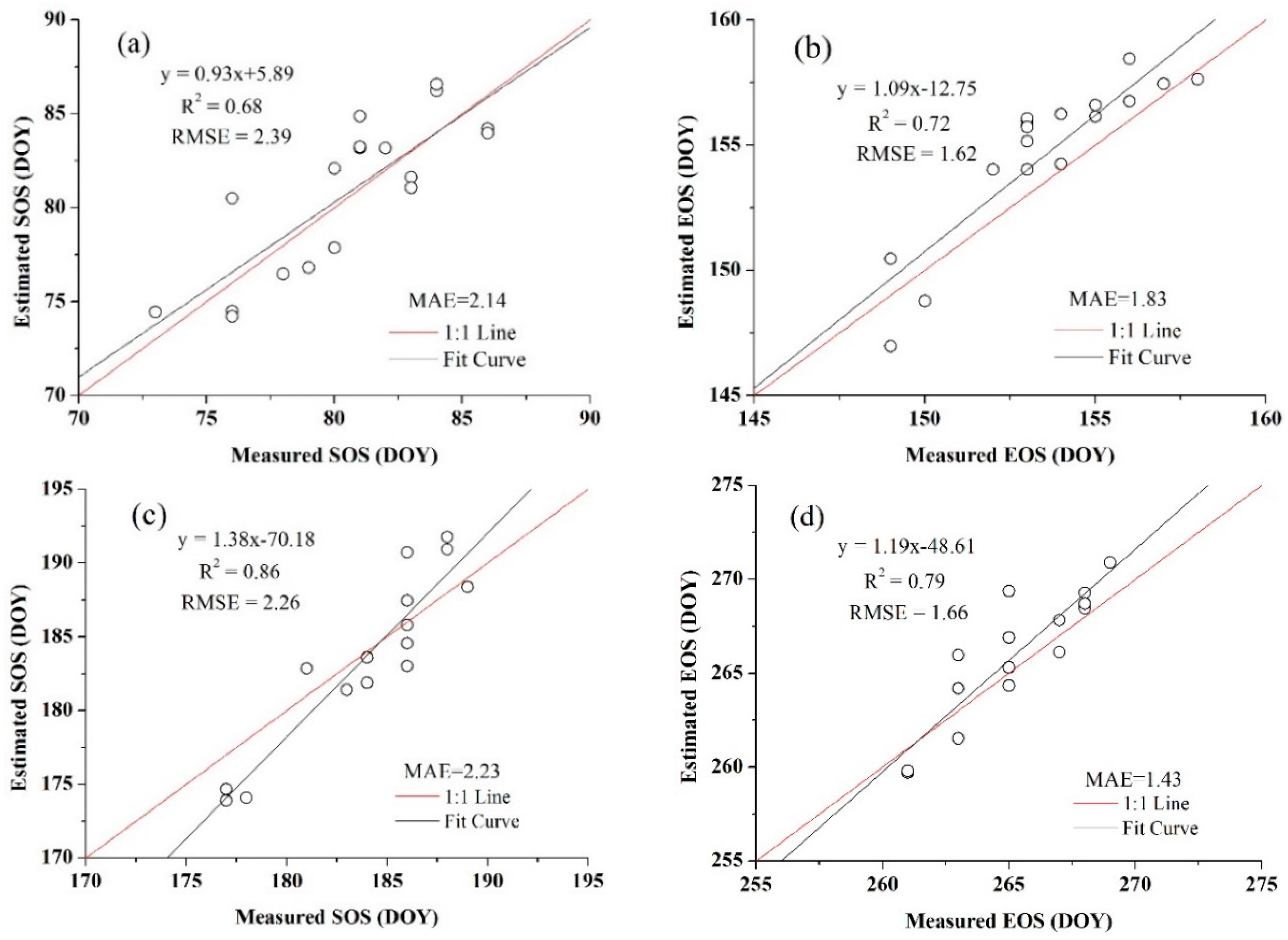
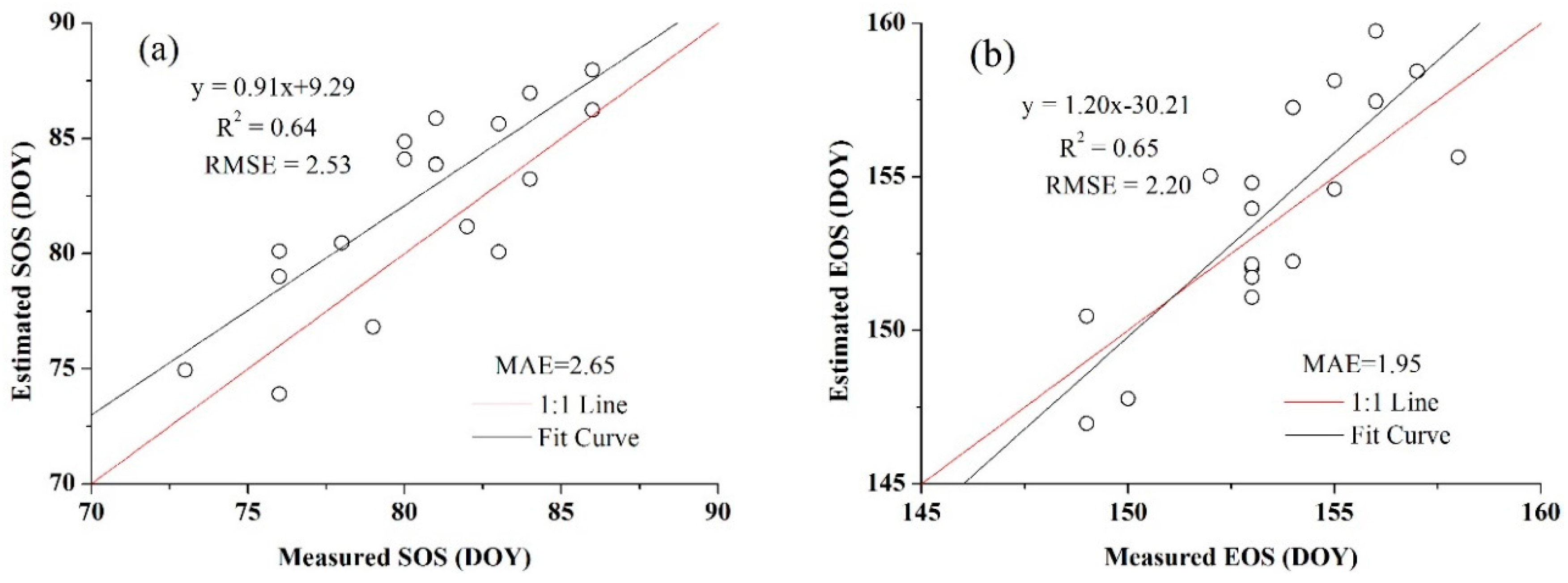
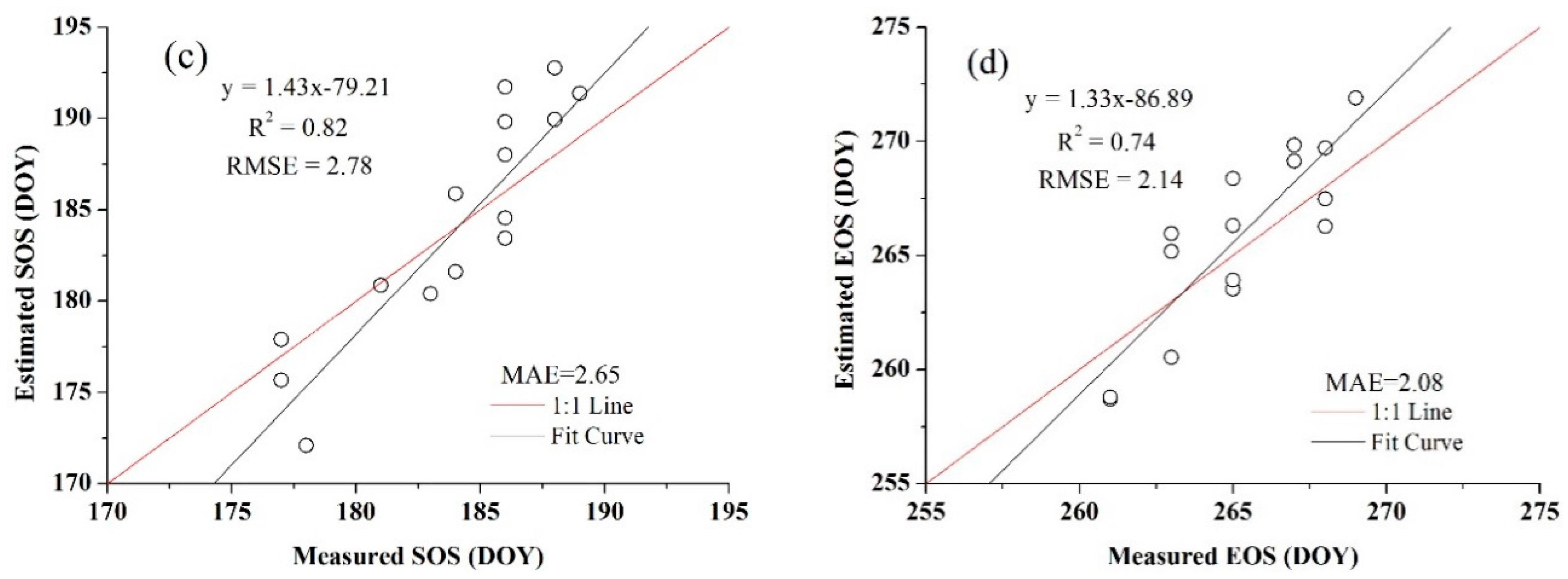
3.4. Accuracy Assessment
4. Results
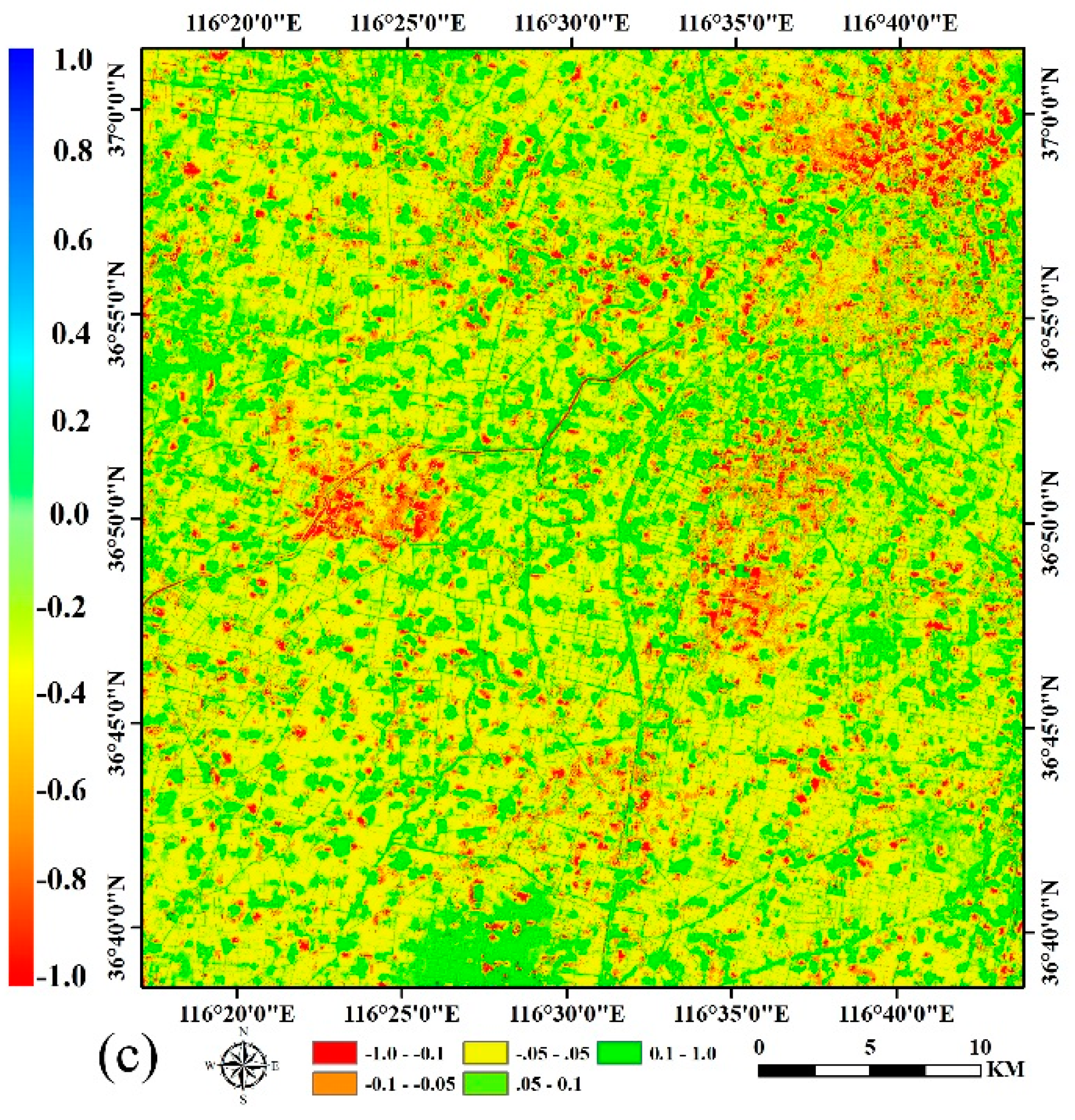
4.1. The STARFM Prediction Results
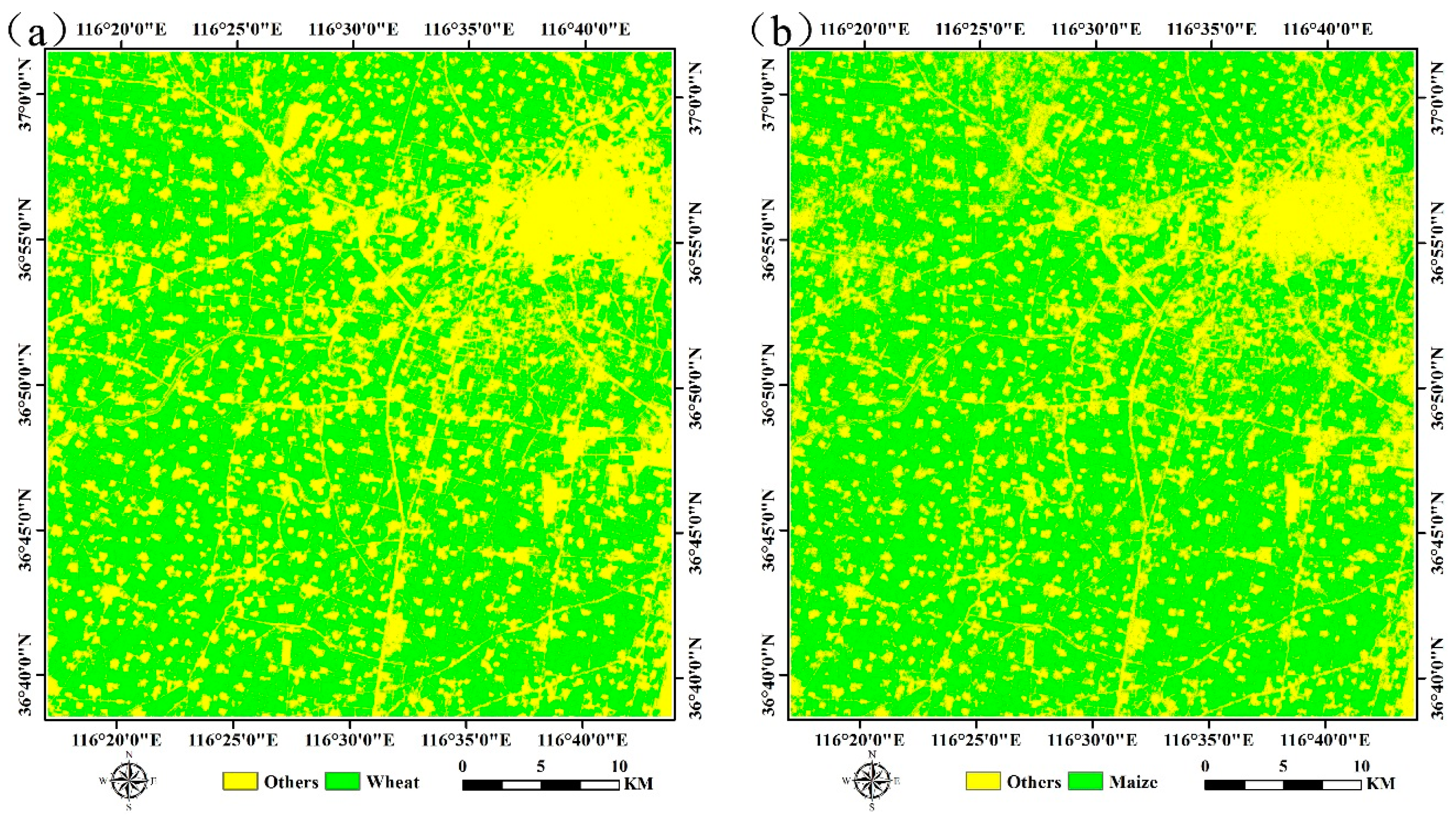
4.2. Crop Classification and Mapping
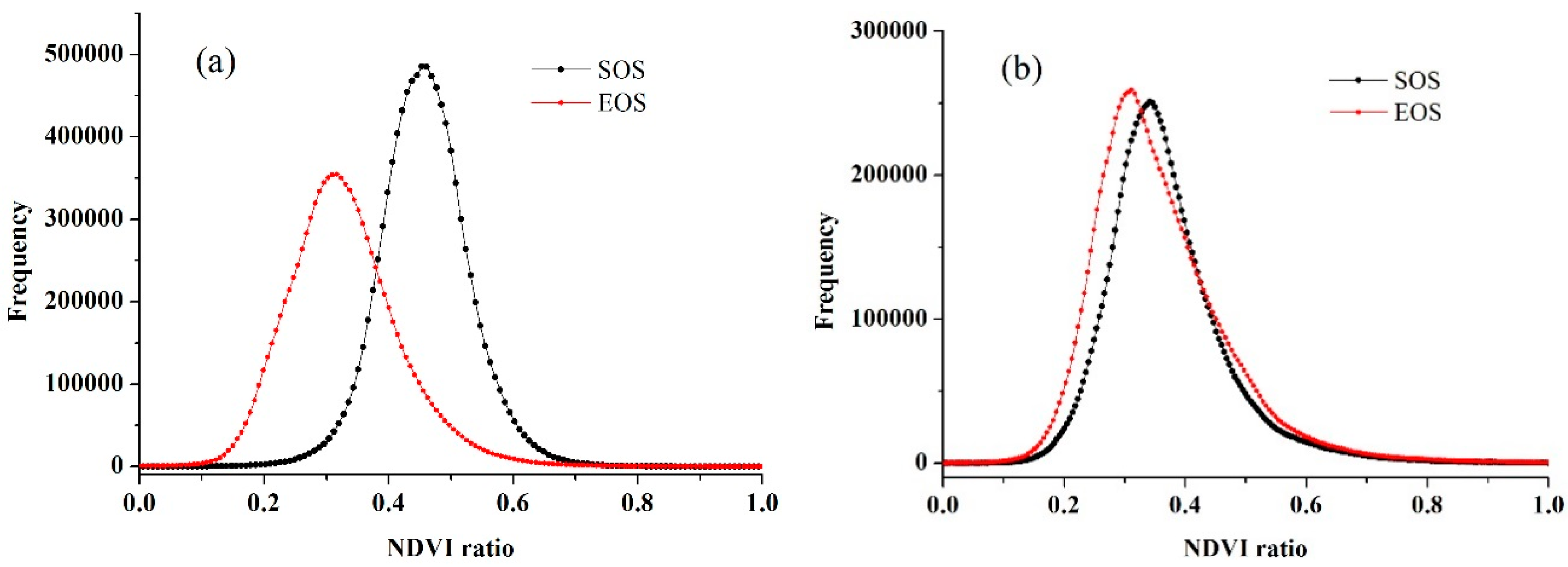
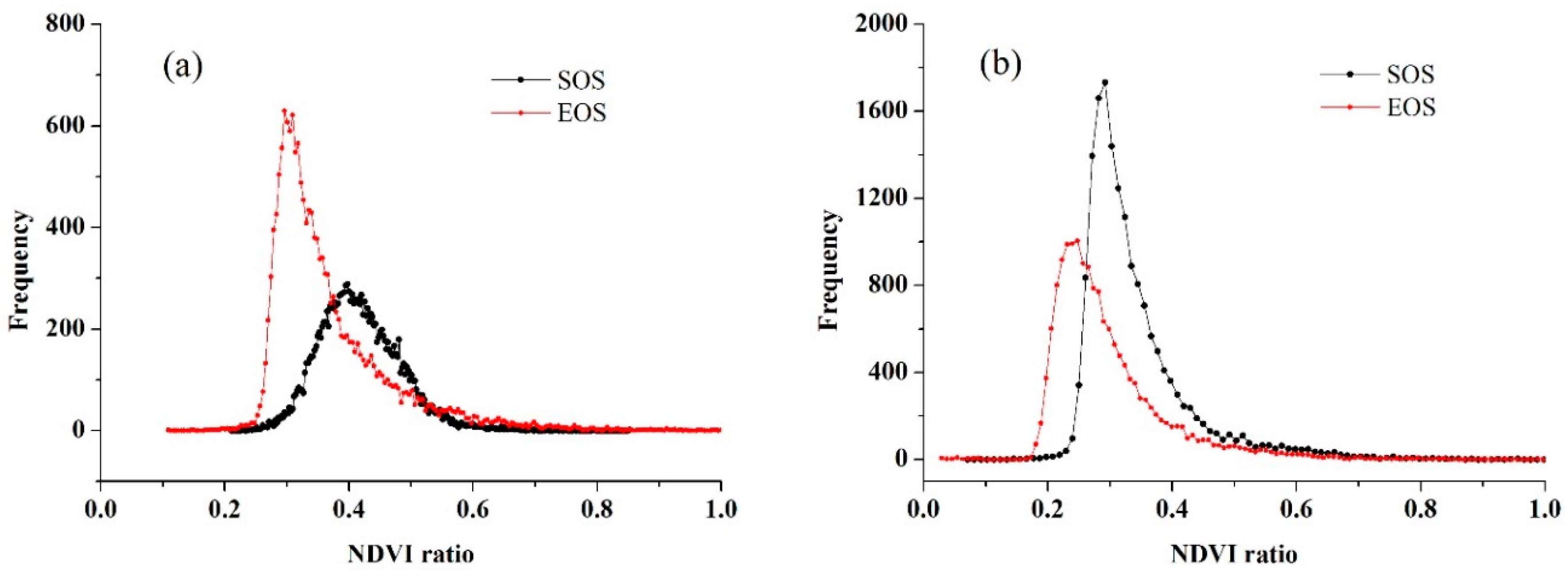
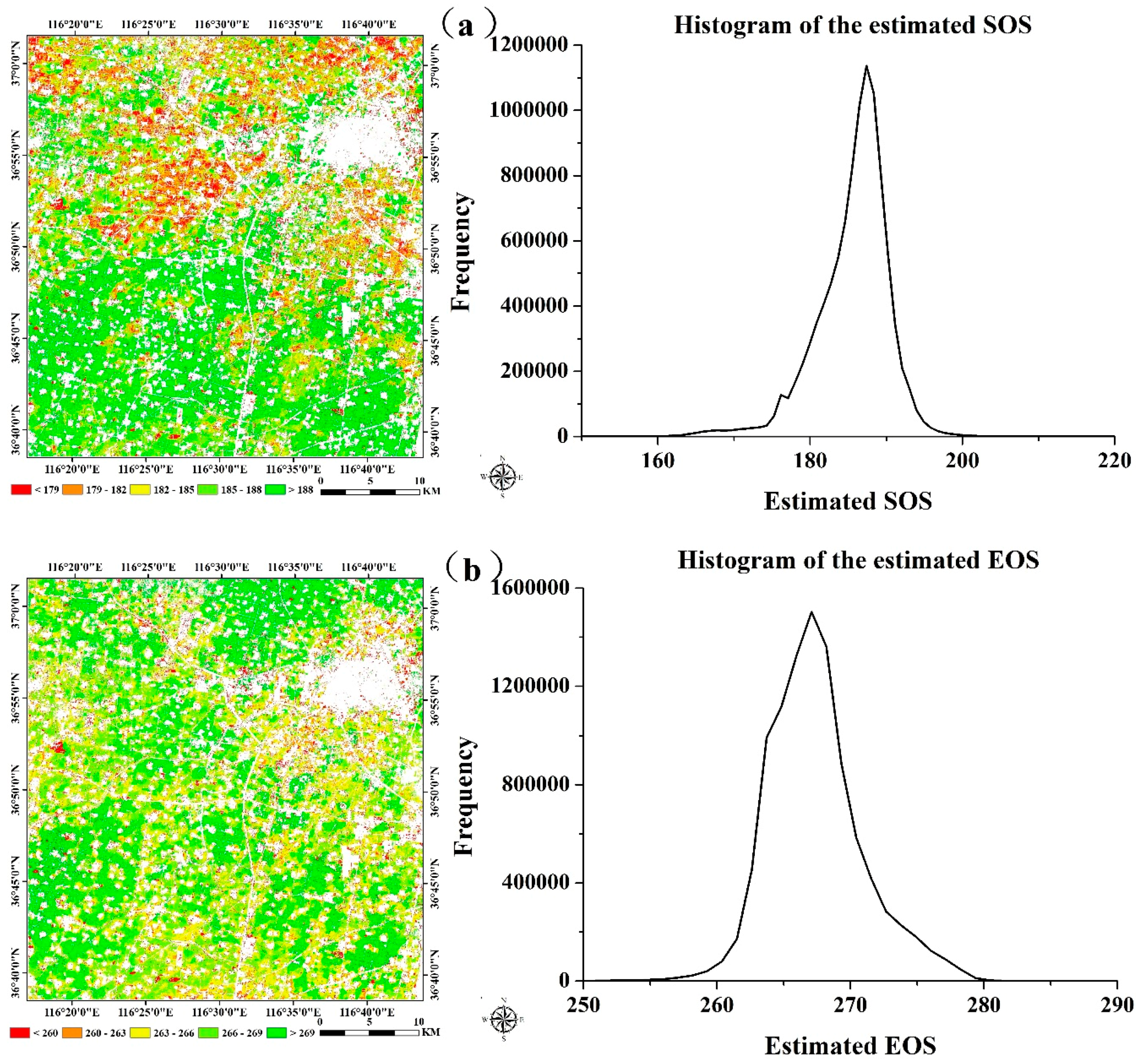
4.3. Crop Phenology Extraction and Mapping
5. Discussion
5.1. High Spatial Resolution Data
5.2. Smoothing Methods
5.3. Data Fusion Algorithms
5.4. Geometric Accuracy and PSF
5.5. VIs’ Selection and Influence
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- White, M.A.; de Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; van Leeuwen, W.J.D.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Begue, A.; Vintrou, E.; Saad, A.; Hiernaux, P. Differences between cropland and rangeland MODIS phenology (start-of-season) in Mali. Int. J. Appl. Earth Obs. Geoinform. 2014, 31, 167–170. [Google Scholar] [CrossRef]
- Soudani, K.; le Maire, G.; Dufrêne, E.; François, C.; Delpierre, N.; Ulrich, E.; Cecchini, S. Evaluation of the onset of green-up in temperate deciduous broadleaf forests derived from Moderate Resolution Imaging Spectroradiometer (MODIS) data. Remote Sens. Environ. 2008, 112, 2643–2655. [Google Scholar] [CrossRef]
- You, X.; Meng, J.; Zhang, M.; Dong, T. Remote sensing based detection of crop phenology for agricultural zones in China using a new threshold method. Remote Sens. 2013, 5, 3190–3211. [Google Scholar] [CrossRef]
- Sakamoto, T.; Wardlow, B.D.; Gitelson, A.A.; Verma, S.B.; Suyker, A.E.; Arkebauer, T.J. A two-step filtering approach for detecting maize and soybean phenology with time-series MODIS data. Remote Sens. Environ. 2010, 114, 2146–2159. [Google Scholar] [CrossRef]
- Zhang, X.; Friedla, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Atzberger, C. Advances in remote sensing of agriculture: Context description, existing operational monitoring systems and major information needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef]
- Sakamoto, T.; Yokozawa, M.; Toritani, H.; Shibayama, M.; Ishitsuka, N.; Ohno, H. A crop phenology detection method using time-series MODIS data. Remote Sens. Environ. 2005, 96, 366–374. [Google Scholar] [CrossRef]
- Cong, N.; Piao, S.; Chen, A.; Wang, X.; Lin, X.; Chen, S.; Han, S.; Zhou, G.; Zhang, X. Spring vegetation green-up date in China inferred from SPOT NDVI data: A multiple model analysis. Agric. For. Meteorol. 2012, 165, 104–113. [Google Scholar] [CrossRef]
- Lu, L.; Wang, C.; Guo, H.; Li, Q. Detecting winter wheat phenology with SPOT-VEGETATION data in the North China Plain. Geocarto Int. 2013, 29, 244–255. [Google Scholar] [CrossRef]
- Brown, M.E.; de Beurs, K.M.; Marshall, M. Global phenological response to climate change in crop areas using satellite remote sensing of vegetation, humidity and temperature over 26 years. Remote Sens. Environ. 2012, 126, 174–183. [Google Scholar] [CrossRef]
- Heumann, B.W.; Seaquist, J.W.; Eklundh, L.; Jönsson, P. AVHRR derived phenological change in the Sahel and Soudan, Africa, 1982–2005. Remote Sens. Environ. 2007, 108, 385–392. [Google Scholar] [CrossRef]
- Zhang, M.; Zhou, Q.; Chen, Z.; Liu, J.; Zhou, Y.; Cai, C. Crop discrimination in Northern China with double cropping systems using fourier analysis of time-series MODIS data. Int. J. Appl. Earth Obs. Geoinform. 2008, 10, 476–485. [Google Scholar]
- Pan, Z.; Huang, J.; Zhou, Q.; Wang, L.; Cheng, Y.; Zhang, H.; Blackburn, G.A.; Yan, J.; Liu, J. Mapping crop phenology using NDVI time-series derived from HJ-1 A/B data. Int. J. Appl. Earth Obs. Geoinform. 2015, 34, 188–197. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the Landsat and MODIS surface reflectance: Predicting daily landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Seitz, N.; White, J.C.; Gao, F.; Masek, J.G.; Stenhouse, G. Generation of dense time series synthetic Landsat data through data blending with MODIS using a spatial and temporal adaptive reflectance fusion model. Remote Sens. Environ. 2009, 113, 1988–1999. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Gao, F.; Chen, X.; Masek, J.G. An enhanced spatial and temporal adaptive reflectance fusion model for complex heterogeneous regions. Remote Sens. Environ. 2010, 114, 2610–2623. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, L.; Xie, D.; Yin, X.; Liu, C.; Liu, G. Application of synthetic NDVI time series blended from Landsat and MODIS data for grassland biomass estimation. Remote Sens. 2016, 8, 10. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, M.; Zhang, X.; Zeng, H.; Wu, B. Mapping winter wheat biomass and yield using time series data blended from PROBA-V 100- and 300-m S1 products. Remote Sens. 2016, 8, 824. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, X.; Liu, D. Blending MODIS and Landsat images for urban flood mapping. Int. J. Remote Sens. 2014, 35, 3237–3253. [Google Scholar] [CrossRef]
- Liu, H.; Weng, Q. Enhancing temporal resolution of satellite imagery for public health studies: A case study of West Nile Virus outbreak in Los Angeles in 2007. Remote Sens. Environ. 2012, 117, 57–71. [Google Scholar] [CrossRef]
- Tewes, A.; Thonfeld, F.; Schmidt, M.; Oomen, R.; Zhu, X.; Dubovyk, O.; Menz, G.; Schellberg, J. Using RapidEye and MODIS data fusion to monitor vegetation dynamics in semi-arid rangelands in South Africa. Remote Sens. 2015, 7, 6510–6534. [Google Scholar] [CrossRef]
- Liu, X.; Bo, Y.; Zhang, J.; He, Y. Classification of C3 and C4 vegetation types using MODIS and ETM+ blended high spatio-temporal resolution data. Remote Sens. 2015, 7, 15244–15268. [Google Scholar] [CrossRef]
- Singha, M.; Wu, B.; Zhang, M. An object-based paddy rice classification using multi-spectral data and crop phenology in Assam, northeast India. Remote Sens. 2016, 8, 479. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, B.; Meng, J. Quantifying winter wheat residue biomass with a spectral angle index derived from China Environmental Satellite data. Int. J. Appl. Earth Obs. Geoinform. 2014, 32, 105–113. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, L.; Yan, C.; Wang, Z. ChinaCover 2010: Methodology and features. In Proceedings of the GeoInformatics, Hong Kong, China, 15–17 June 2012.
- Zhang, M.; Wu, B.; Yu, M.; Zou, W.; Zheng, Y. Crop condition assessment with adjusted NDVI using the uncropped arable land ratio. Remote Sens. 2014, 6, 5774–5794. [Google Scholar] [CrossRef]
- Tian, F.; Wang, Y.; Fensholt, R.; Wang, K.; Zhang, L.; Huang, Y. Mapping and evaluation of NDVI trends from synthetic time series obtained by blending Landsat and MODIS data around a Coalfield on the Loess Plateau. Remote Sens. 2013, 5, 4255–4279. [Google Scholar] [CrossRef]
- Jarihani, A.; McVicar, T.; Van Niel, T.; Emelyanova, I.; Callow, J.; Johansen, K. Blending Landsat and MODIS data to generate multispectral indices: A comparison of “index-then-blend” and “blend-then-index” approaches. Remote Sens. 2014, 6, 9213–9238. [Google Scholar] [CrossRef] [Green Version]
- Jönsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- The Information of SPOT5 Take5 Product Format. Available online: http://www.cesbio.ups-tlse.fr/multitemp/?page_id=1822 (accessed on 11 March 2016).
- Lara, B.; Gandini, M. Assessing the performance of smoothing functions to estimate land surface phenology on temperate grassland. Int. J. Remote Sens. 2016, 37, 1801–1813. [Google Scholar] [CrossRef]
- Lu, L.; Guo, H. Extraction method of winter wheat phenology from time series of SPOT/VEGETATION data. Trans. CSAE 2009, 25, 174–179. (In Chinese) [Google Scholar]
- Lloyd, D. A phenological classification of terrestrial vegetation cover using shortwave vegetation index imagery. Int. J. Remote Sens. 1990, 11, 2269–2279. [Google Scholar] [CrossRef]
- Pan, Y.; Li, L.; Zhang, J.; Liang, S.; Zhu, X.; Sulla-Menashe, D. Winter wheat area estimation from MODIS-EVI time series data using the crop proportion phenology index. Remote Sens. Environ. 2012, 119, 232–242. [Google Scholar] [CrossRef]
- Jönsson, P.; Eklundh, L. Timesat-a program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef]
- Shao, J. Linear model selection by cross-validation. J. Am. Stat. Assoc. 1993, 88, 486–494. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First experience with Sentinel-2 data for crop and tree species classifications in central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Marie-Julie, L.; François, W.; Defourny, P. Cropland mapping over Sahelian and Sudanian agrosystems: A knowledge-based approach using PROBA-V time series at 100-m. Remote Sens. 2016, 8, 232–254. [Google Scholar]
- Atzberger, C.; Eilers, P.H.C. Evaluating the effectiveness of smoothing algorithms in the absence of ground reference measurements. Int. J. Remote Sens. 2011, 32, 3689–3709. [Google Scholar] [CrossRef]
- Knauer, K.; Gessner, U.; Fensholt, R.; Kuenzer, C. An ESTARFM fusion framework for the generation of large-scale time series in cloud-prone and heterogeneous landscapes. Remote Sens. 2016, 8, 425. [Google Scholar] [CrossRef]
- Kaiser, G.; Schneider, W. Estimation of sensor point spread function by spatial subpixel analysis. Int. J. Remote Sens. 2008, 29, 2137–2155. [Google Scholar] [CrossRef]
- Duveiller, G.; Baret, F.; Defourny, P. Crop specific green area index retrieval from modis data at regional scale by controlling pixel-target adequacy. Remote Sens. Environ. 2011, 115, 2686–2701. [Google Scholar] [CrossRef]
- Zurita-Milla, R.; Kaiser, G.; Clevers, J.G.P.W.; Schneider, W.; Schaepman, M.E. Downscaling time series of meris full resolution data to monitor vegetation seasonal dynamics. Remote Sens. Environ. 2009, 113, 1874–1885. [Google Scholar] [CrossRef]
- Schowengerdt, R.A. Remote Sensing: Models and Methods for Image Processing, 2nd ed.; Academic Press: San Diego, CA, USA, 1997; pp. 67–83. [Google Scholar]
- Duveiller, G.; Defourny, P. A conceptual framework to define the spatial resolution requirements for agricultural monitoring using remote sensing. Remote Sens. Environ. 2010, 114, 2637–2650. [Google Scholar] [CrossRef]
- Zurita-Milla, R.; Clevers, J.G.P.W.; Schaepman, M.E.; Kneubuehler, M. Effects of MERIS l1b radiometric calibration on regional land cover mapping and land products. Int. J. Remote Sens. 2007, 28, 653–673. [Google Scholar] [CrossRef]
- Tan, B.; Woodcock, C.E.; Hu, J.; Zhang, P.; Ozdogan, M.; Huang, D.; Yang, W.; Knyazikhin, Y.; Myneni, R.B. The impact of gridding artifacts on the local spatial properties of modis data: Implications for validation, compositing, and band-to-band registration across resolutions. Remote Sens. Environ. 2006, 105, 98–114. [Google Scholar] [CrossRef]
- Kross, A.; McNairn, H.; Lapen, D.; Sunohara, M.; Champagne, C. Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. Int. J. Appl. Earth Obs. Geoinform. 2015, 34, 235–248. [Google Scholar] [CrossRef]
- Gao, S.; Niu, Z.; Huang, N.; Hou, X. Estimating the leaf area index, height and biomass of maize using HJ-1 and RADARSAT-2. Int. J. Appl. Earth Obs. Geoinform. 2013, 24, 1–8. [Google Scholar] [CrossRef]
- Jin, X.; Yang, G.; Xu, X.; Yang, H.; Feng, H.; Li, Z.; Shen, J.; Lan, Y.; Zhao, C. Combined multi-temporal optical and radar parameters for estimating LAI and biomass in winter wheat using HJ and RADARSAT-2 data. Remote Sens. 2015, 7, 13251–13272. [Google Scholar] [CrossRef]
- Meng, J.; Xu, J.; You, X. Optimizing soybean harvest date using HJ-1 satellite imagery. Precis. Agric. 2015, 16, 164–179. [Google Scholar] [CrossRef]



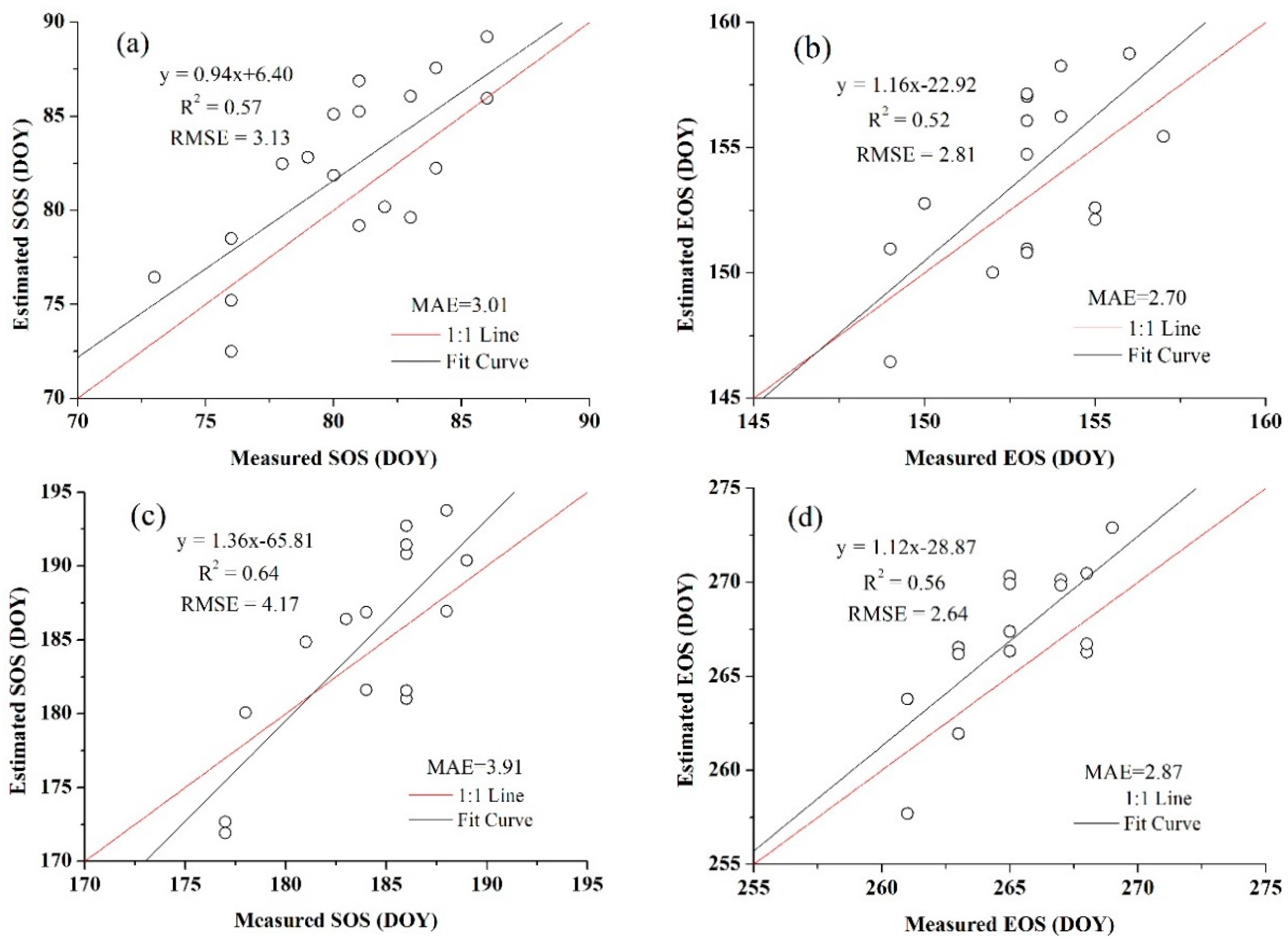
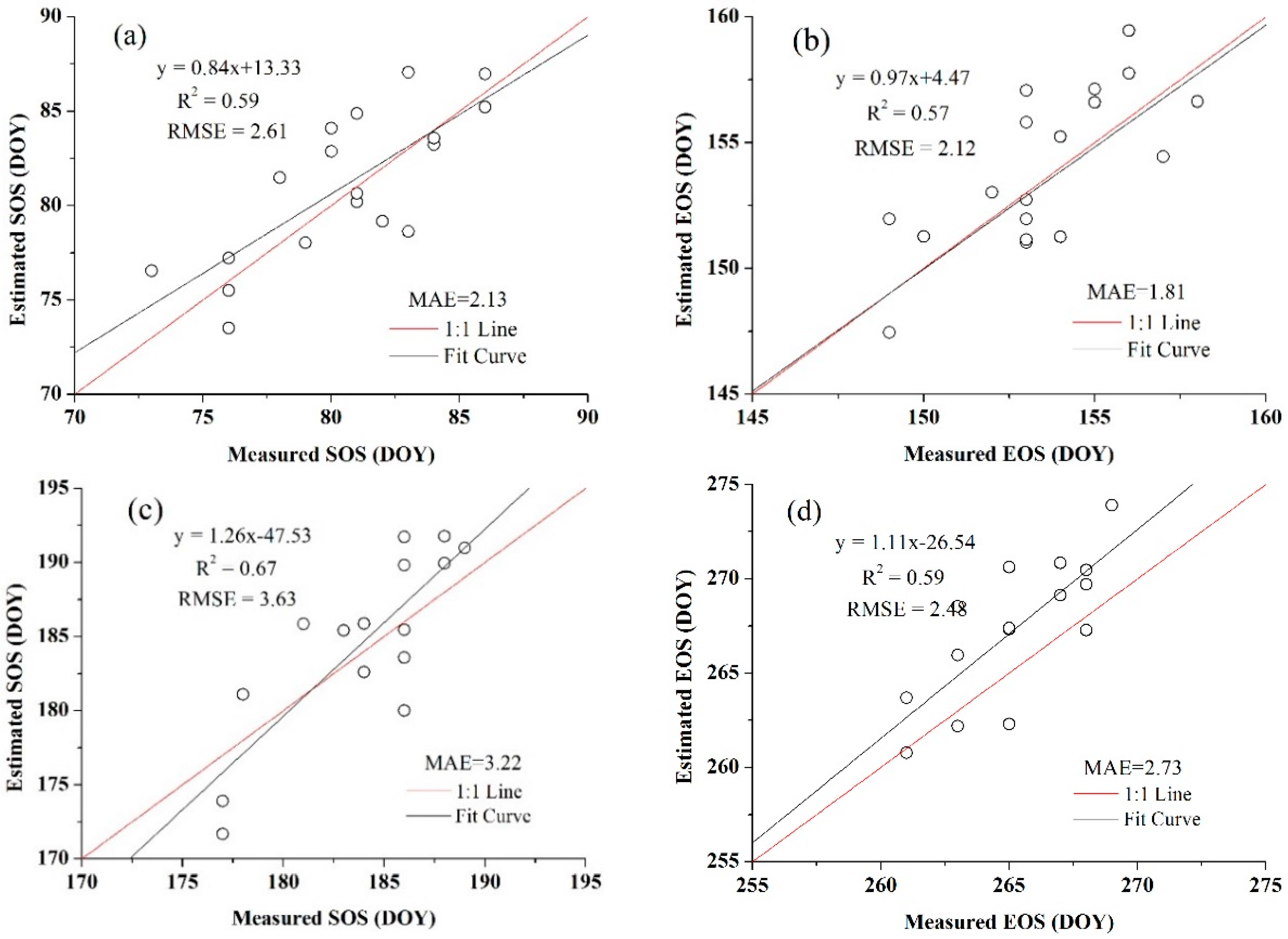
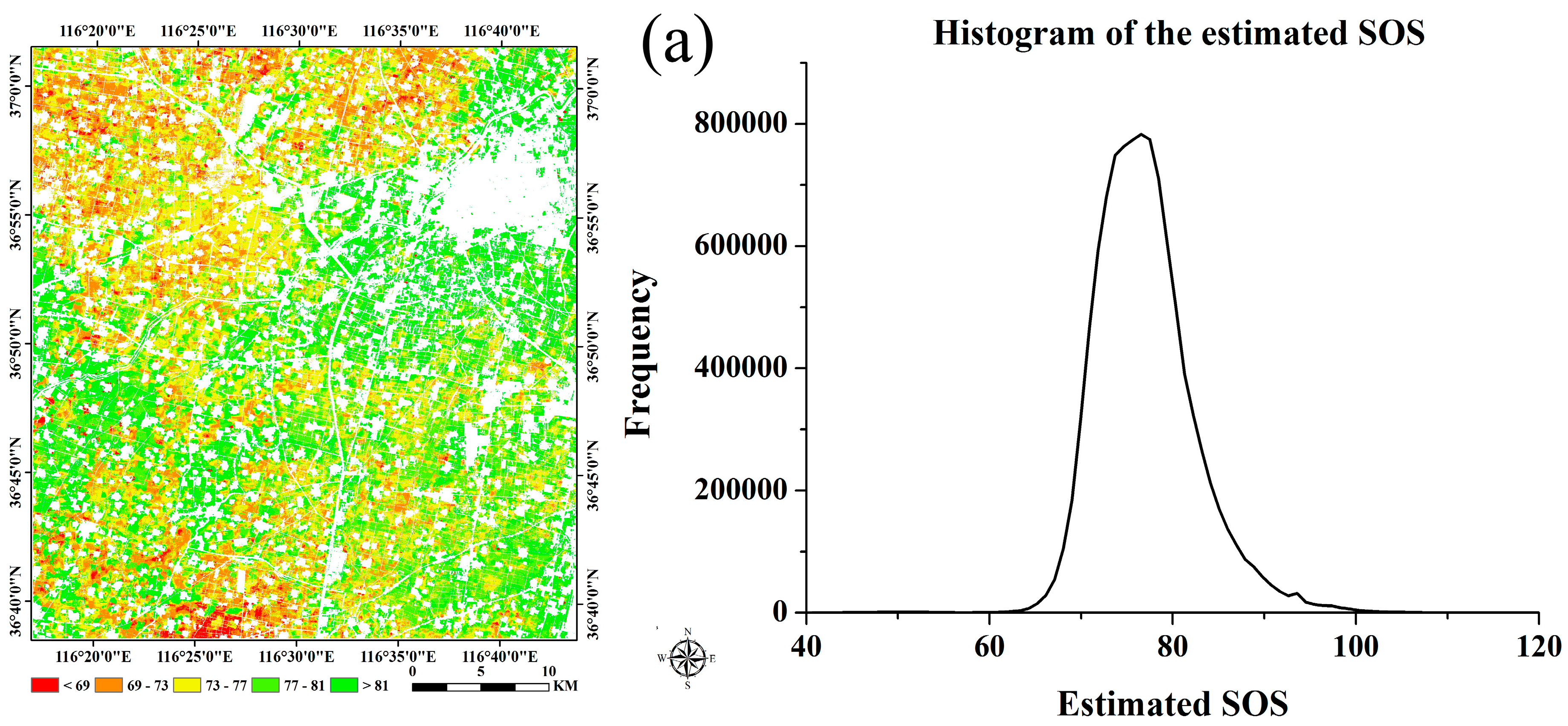
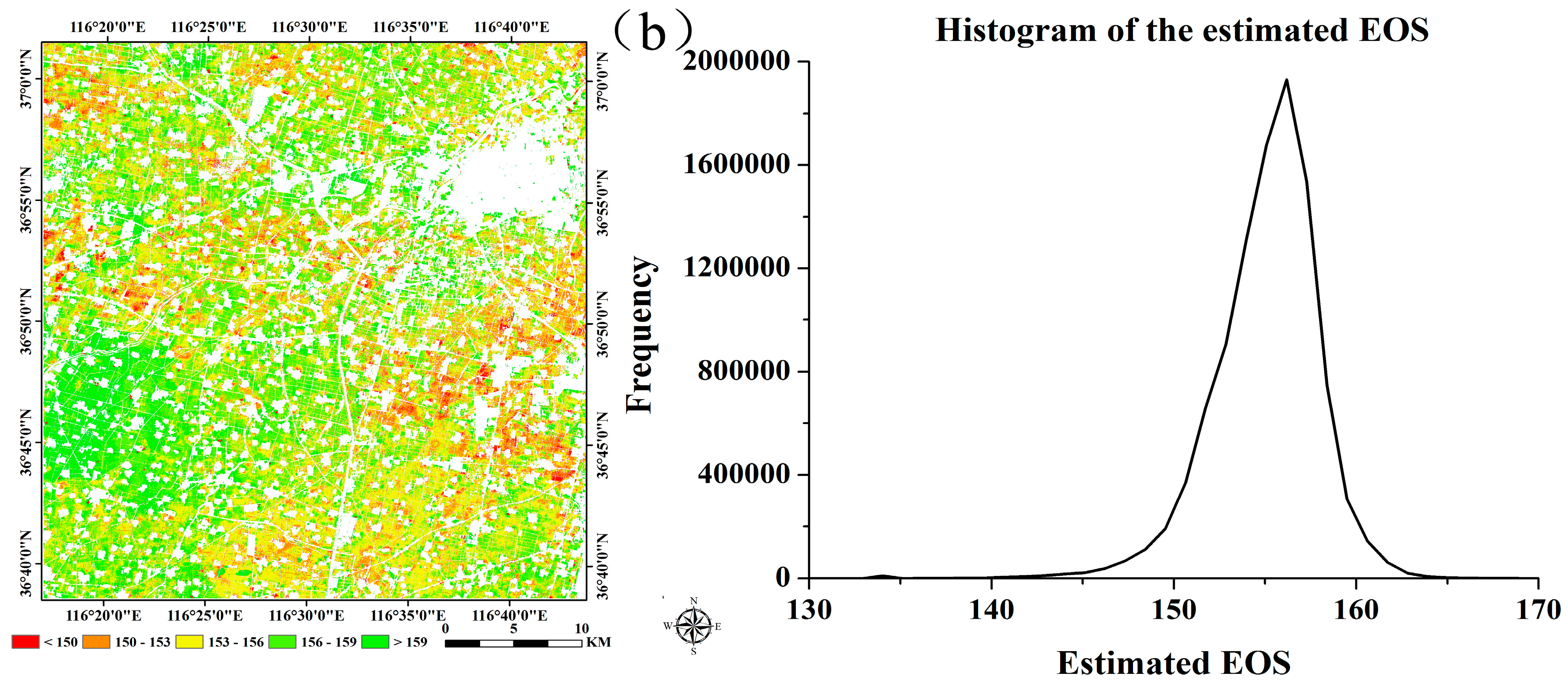
| Winter Wheat | Summer Maize | ||
|---|---|---|---|
| Phenology Stage | Date (DOY) | Phenology Stage | Date (DOY) |
| Sowing | 288 | Sowing | 163 |
| Emergence | 298 | Emergence | 169 |
| Tillering | 325 | Seven leaf | 186 (SOS) |
| Wintering | 349–51 (next year) | Tasseling | 222 |
| Jointing | 81 (SOS) | Silking | 227 |
| Booting | 103 | Maturity | 265 (EOS) |
| Maturity | 153 (EOS) | Harvest | 274 |
| Harvest | 160 | - | - |
| Wheat Season | Maize Season | ||
|---|---|---|---|
| Acquisition Date (Month/Day) | DOY | Acquisition Date (Month/Day) | DOY |
| 4/23 | 113 | 7/2 | 183 |
| 5/8 | 128 | 8/11 | 223 |
| 5/13 | 133 | 8/16 | 228 |
| 5/23 | 143 | 8/21 | 233 |
| 5/28 | 148 | 9/15 | 258 |
| Class | SPOT5 Data | MODIS Data | ||
|---|---|---|---|---|
| Producer’s Accuracy | User’s Accuracy | Producer’s Accuracy | User’s Accuracy | |
| Wheat | 89.13% | 83.67% | 73.91% | 67.33% |
| Others | 84.62% | 89.79% | 68.26% | 74.74% |
| Overall Accuracy: 86.73%; Kappa: 0.7347 | Overall Accuracy: 70.92%; Kappa: 0.4210 | |||
| Class | SPOT5 Data | MODIS Data | ||
|---|---|---|---|---|
| Producer’s Accuracy | User’s Accuracy | Producer’s Accuracy | User’s Accuracy | |
| Maize | 91.26% | 87.85% | 75.73% | 70.91% |
| Others | 89.43% | 92.44% | 73.98% | 78.45% |
| Overall Accuracy: 90.27%; Kappa: 0.8044 | Overall Accuracy: 74.78%; Kappa: 0.4944 | |||
| Filtering Methods | Crop Types | |
|---|---|---|
| Winter Wheat | Summer Maize | |
| A-G | 0.0158 | 0.0291 |
| D-L | 0.0235 | 0.0453 |
| S-G | 0.0152 | 0.0266 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Wu, B.; Zhang, M.; Zeng, H. Crop Phenology Detection Using High Spatio-Temporal Resolution Data Fused from SPOT5 and MODIS Products. Sensors 2016, 16, 2099. https://doi.org/10.3390/s16122099
Zheng Y, Wu B, Zhang M, Zeng H. Crop Phenology Detection Using High Spatio-Temporal Resolution Data Fused from SPOT5 and MODIS Products. Sensors. 2016; 16(12):2099. https://doi.org/10.3390/s16122099
Chicago/Turabian StyleZheng, Yang, Bingfang Wu, Miao Zhang, and Hongwei Zeng. 2016. "Crop Phenology Detection Using High Spatio-Temporal Resolution Data Fused from SPOT5 and MODIS Products" Sensors 16, no. 12: 2099. https://doi.org/10.3390/s16122099






