Phylogeography of Rattus norvegicus in the South Atlantic Ocean
Abstract
:1. Introduction
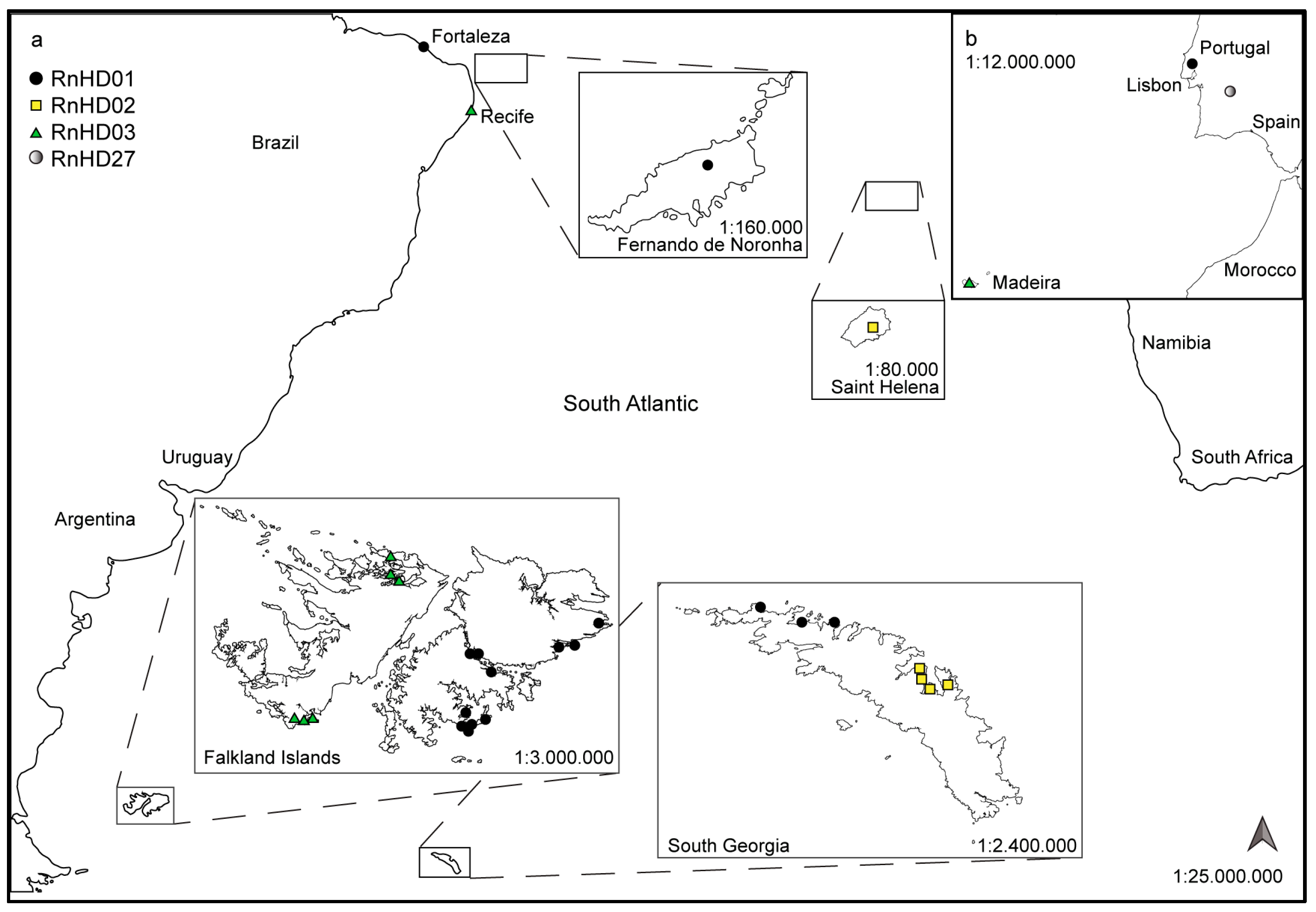
2. Materials and Methods
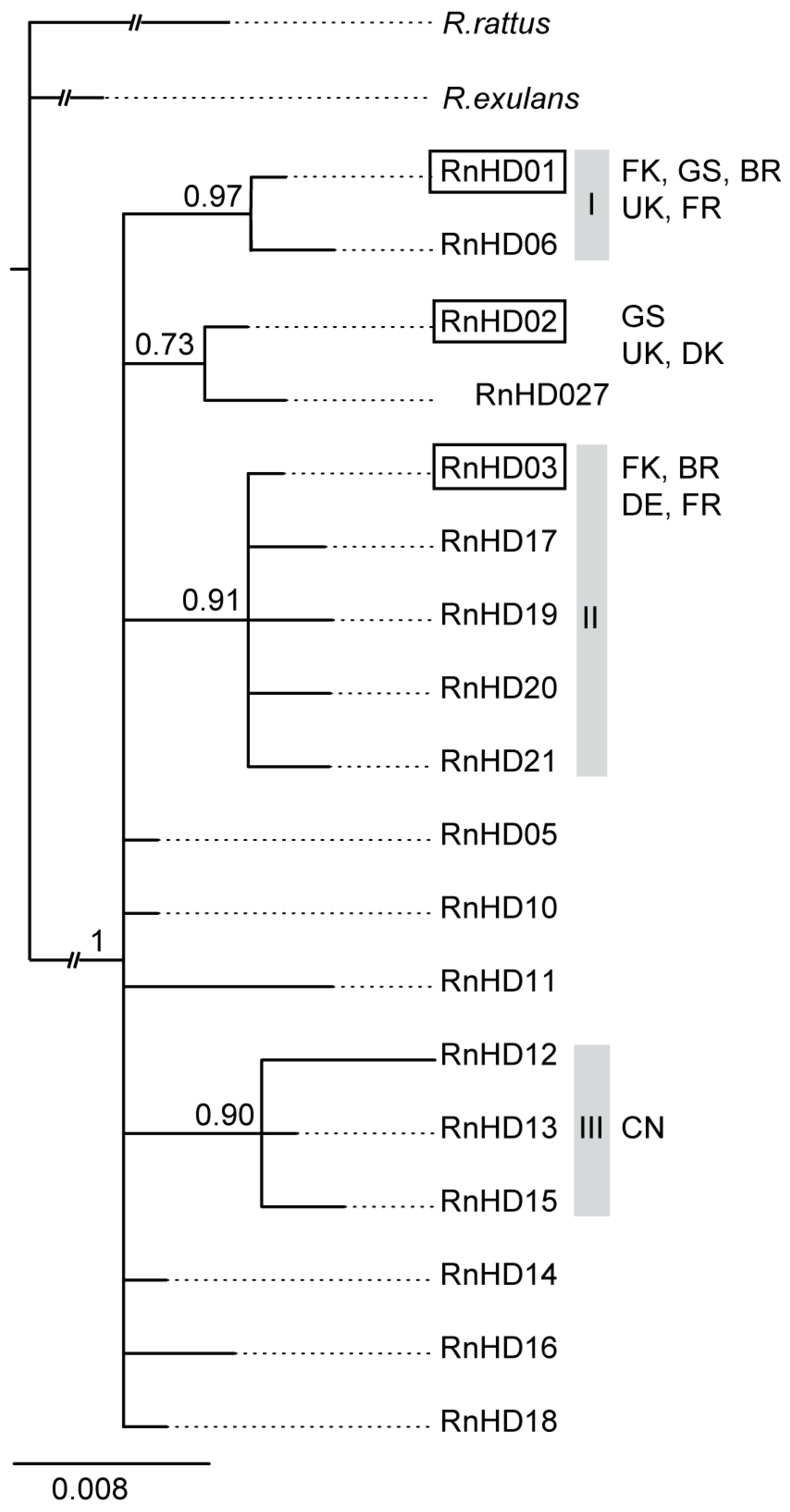
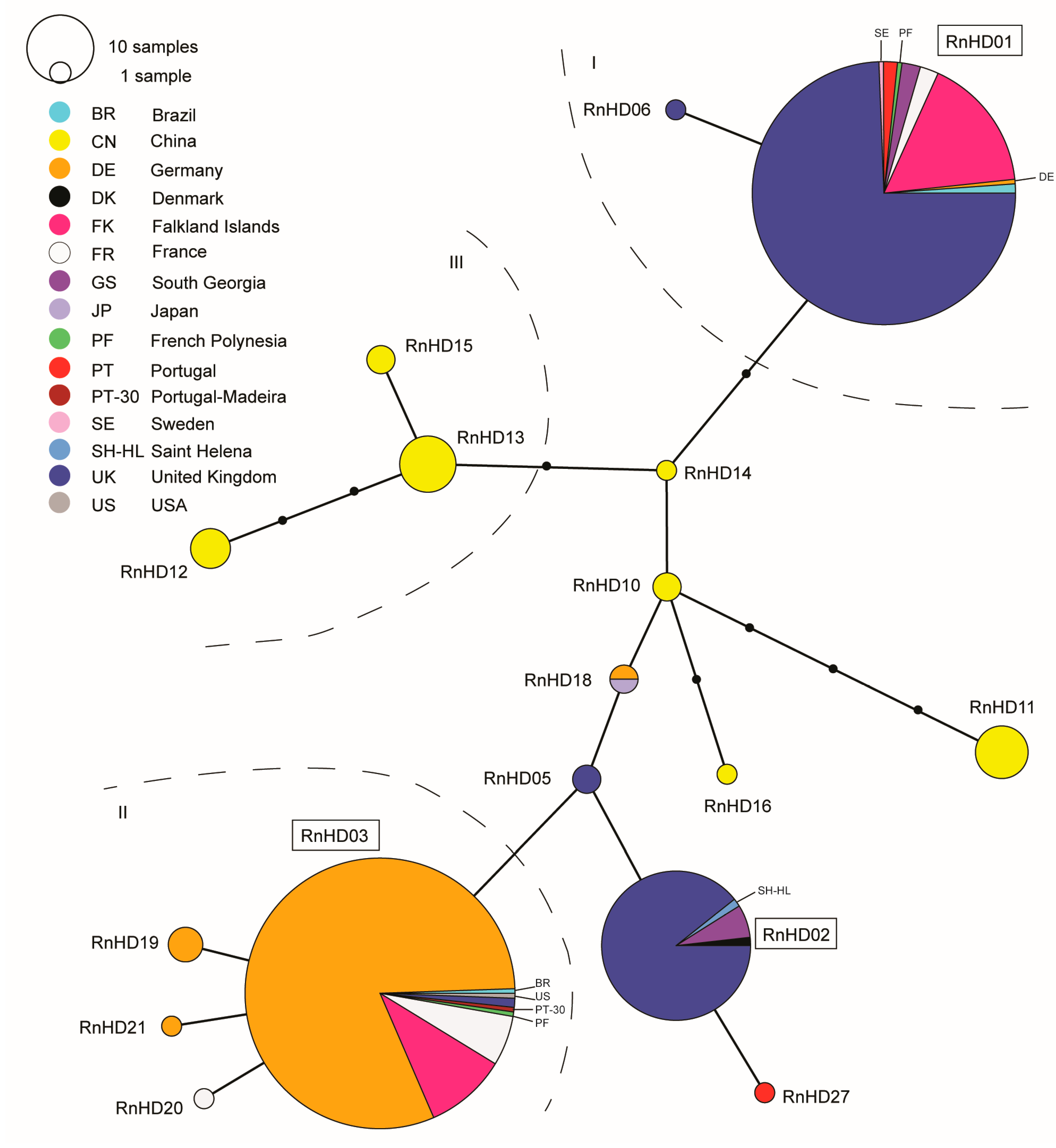
3. Results
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Aplin, K.P.; Chesser, T.; Have, J.T. Evolutionary biology of the genus Rattus: Profile of an archetypal rodent pest. ACIAR Monogr. Ser. 2003, 96, 487–498. [Google Scholar]
- Puckett, E.E.; Park, J.; Combs, M.; Blum, M.J.; Bryant, J.E.; Caccone, A.; Costa, F.; Deinum, E.E.; Esther, A.; Himsworth, C.G.; et al. Global population divergence and admixture of the brown rat Rattus norvegicus. Proc. R. Soc. B Biol. Sci. 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, I.A.E. The spread of commensal species of Rattus to oceanic islands and their effects on island avifaunas. In Conservation of Island Birds; ICBP Technical Publication No. 3; Moors, P.J., Ed.; International Council for Bird Preservation: Cambridge, UK, 1985; pp. 35–81. [Google Scholar]
- Dutton, J. Introduced mammals in São Tomé and Príncipe: Possible threats to biodiversity. Biodivers. Conserv. 1994, 3, 927–938. [Google Scholar] [CrossRef]
- Jones, E.P.; Eager, H.M.; Gabriel, S.I.; Jóhannesdóttir, F.; Searle, J.B. Genetic tracking of mice and other bioproxies to infer human history. Trends Genet. 2013, 29, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Robertson, B.C.; Gemmell, N.J. Defining eradication units to control invasive pests. J. Appl. Ecol. 2004, 41, 1042–1048. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Pascal, M.; Samadi, S. Establishing causes of eradication failure based on genetics: Case study of ship rat eradication in Ste. Anne archipelago. Conserv. Biol. 2007, 21, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Piertney, S.B.; Black, A.; Watt, L.; Christie, D.; Poncet, S.; Collins, M.A. Resolving patterns of population genetic and phylogeographic structure to inform control and eradication initiatives for brown rats Rattus norvegicus on South Georgia. J. Appl. Ecol. 2016, 53, 332–339. [Google Scholar] [CrossRef]
- Matisoo-Smith, E.; Roberts, R.M.; Irwin, G.J.; Allen, J.S.; Penny, D.; Lambert, D.M. Patterns of prehistoric human mobility in Polynesia indicated by mtDNA from the Pacific rat. Proc. Nalt. Acad. Sci. USA 1998, 95, 15145–15150. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.A.; Gullberg, A.; Spotorno, A.E.; Arnason, U.; Janke, A. Radiation of extant marsupials after the K/T boundary: Evidence from complete mitochondrial genomes. J. Mol. Evol. 2003, 57, S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Schlick, N.E.; Jensen-Seaman, M.I.; Orlebeke, K.; Kwitek, A.E.; Jacob, H.J.; Lazar, J. Sequence analysis of the complete mitochondrial DNA in 10 commonly used inbred rat strains. Am. J. Physiol. Cell Physiol. 2006, 291, C1183–C1192. [Google Scholar] [CrossRef] [PubMed]
- Haniza, M.Z.H.; Adams, S.; Jones, E.P.; MacNicoll, A.; Mallon, E.B.; Smith, R.H.; Lambert, M.S. Large-scale structure of brown rat (Rattus norvegicus) populations in England: Effects on rodenticide resistance. PeerJ 2015, 3, e1458. [Google Scholar] [CrossRef] [PubMed]
- Robins, J.H.; Hingston, M.; Matisoo-Smith, E.; Ross, H.A. Identifying Rattus species using mitochondrial DNA. Mol. Ecol. Notes 2007, 7, 717–729. [Google Scholar] [CrossRef]
- Abhyankar, A.; Park, H.-B.; Tonolo, G.; Luthman, H. Comparative sequence analysis of the non-protein-coding mitochondrial DNA of inbred rat strains. PLoS ONE 2009, 4, e8148. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, D.-Y.; Chen, W.; Li, J.-L.; Luo, F.; Li, Q.; Ling, J.-X.; Liu, Y.-Y.; Xiong, H.-R.; Ding, X.-H.; et al. Genetic analysis of hantaviruses and their rodent hosts in central-south China. Virus Res. 2012, 163, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lan, Z.; Kohn, M.H. Mitochondrial DNA phylogeography of the Norway rat. PLoS ONE 2014, 9, e88425. [Google Scholar] [CrossRef] [PubMed]
- Kaleme, P.; Bates, J.; Belesi, H.; Bowie, R.; Gambalemoke, M.; Kerbis-Peterhans, J.; Michaux, J.; Mwanga, J.; Ndara, B.; Taylor, P.; et al. Origin and putative colonization routes for invasive rodent taxa in the Democratic Republic of Congo. Afr. Zool. 2011, 46, 133–145. [Google Scholar] [CrossRef]
- Usdin, K.; Chevret, P.; Catzeflis, F.M.; Verona, R.; Furano, A.V. Ll (LINE-l) retrotransposable elements provide a “fossil” record of the phylogenetic history of murid rodents. Mol. Biol. Evol. 1995, 12, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART v1.7. Available online: http://popart.otago.ac.nz (accessed on 12 July 2016).
- Tabak, M.A.; Poncet, S.; Passfield, K.; Carling, M.D.; Martinez del Rio, C. The relationship between distance and genetic similarity among invasive rat populations in the Falkland Islands. Conserv. Genet. 2015, 16, 125–135. [Google Scholar] [CrossRef]
- Jones, E.P.; Jensen, J.-K.; Magnussen, E.; Gregersen, N.; Hansen, H.S.; Searle, J.B. A molecular characterization of the charismatic Faroe house mouse. Biol. J. Linn. Soc. 2011, 102, 471–482. [Google Scholar] [CrossRef]
- Gabriel, S.I.; Mathias, M.L.; Searle, J.B. Of mice and the “Age of Discovery”: The complex history of colonization of the Azorean archipelago by the house mouse (Mus musculus) as revealed by mitochondrial DNA variation. J. Evol. Biol. 2015, 28, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Iacucci, A.; Colangelo, P.; Gamberi, V.; Mori, E.; Esther, A.; Baert, K.; Leirs, H.; Petit, T.; Ribas Salvador, A.; Aloise, G.; et al. Reconstructing the phylogeography of an invasive species: Tracing invasion routes of Norway rats (Rattus norvegicus) using mtDNA control region. Hystrix Ital. J. Mammol. 2016, 27, 5. [Google Scholar]
- Fraser, C.I.; Banks, S.C.; Waters, J.M. Priority effects can lead to underestimation of dispersal and invasion potential. Biol. Invasions 2015, 17, 1–8. [Google Scholar] [CrossRef]
- Russell, J.C.; Abdelkrim, J.; Fewster, R.M. Early colonisation population structure of a Norway rat island invasion. Biol. Invasions 2009, 11, 1557–1567. [Google Scholar] [CrossRef]
- Russell, J.C.; Towns, D.R.; Anderson, S.H.; Clout, M.N. Intercepting the first rat ashore. Nature 2005, 437, 1107. [Google Scholar] [CrossRef] [PubMed]
- Förster, D.W.; Gündüz, İ.; Nunes, A.C.; Gabriel, S.; Ramalhinho, M.G.; Mathias, M.L.; Britton-Davidian, J.; Searle, J.B. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol. Ecol. 2009, 18, 4477–4494. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hingston, M.; Poncet, S.; Passfield, K.; Tabak, M.A.; Gabriel, S.I.; Piertney, S.B.; Russell, J.C. Phylogeography of Rattus norvegicus in the South Atlantic Ocean. Diversity 2016, 8, 32. https://doi.org/10.3390/d8040032
Hingston M, Poncet S, Passfield K, Tabak MA, Gabriel SI, Piertney SB, Russell JC. Phylogeography of Rattus norvegicus in the South Atlantic Ocean. Diversity. 2016; 8(4):32. https://doi.org/10.3390/d8040032
Chicago/Turabian StyleHingston, Melanie, Sally Poncet, Ken Passfield, Michael A. Tabak, Sofia I. Gabriel, Stuart B. Piertney, and James C. Russell. 2016. "Phylogeography of Rattus norvegicus in the South Atlantic Ocean" Diversity 8, no. 4: 32. https://doi.org/10.3390/d8040032





