Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle
Abstract
:1. Introduction
2. Experimental Section
3. Results and Discussion
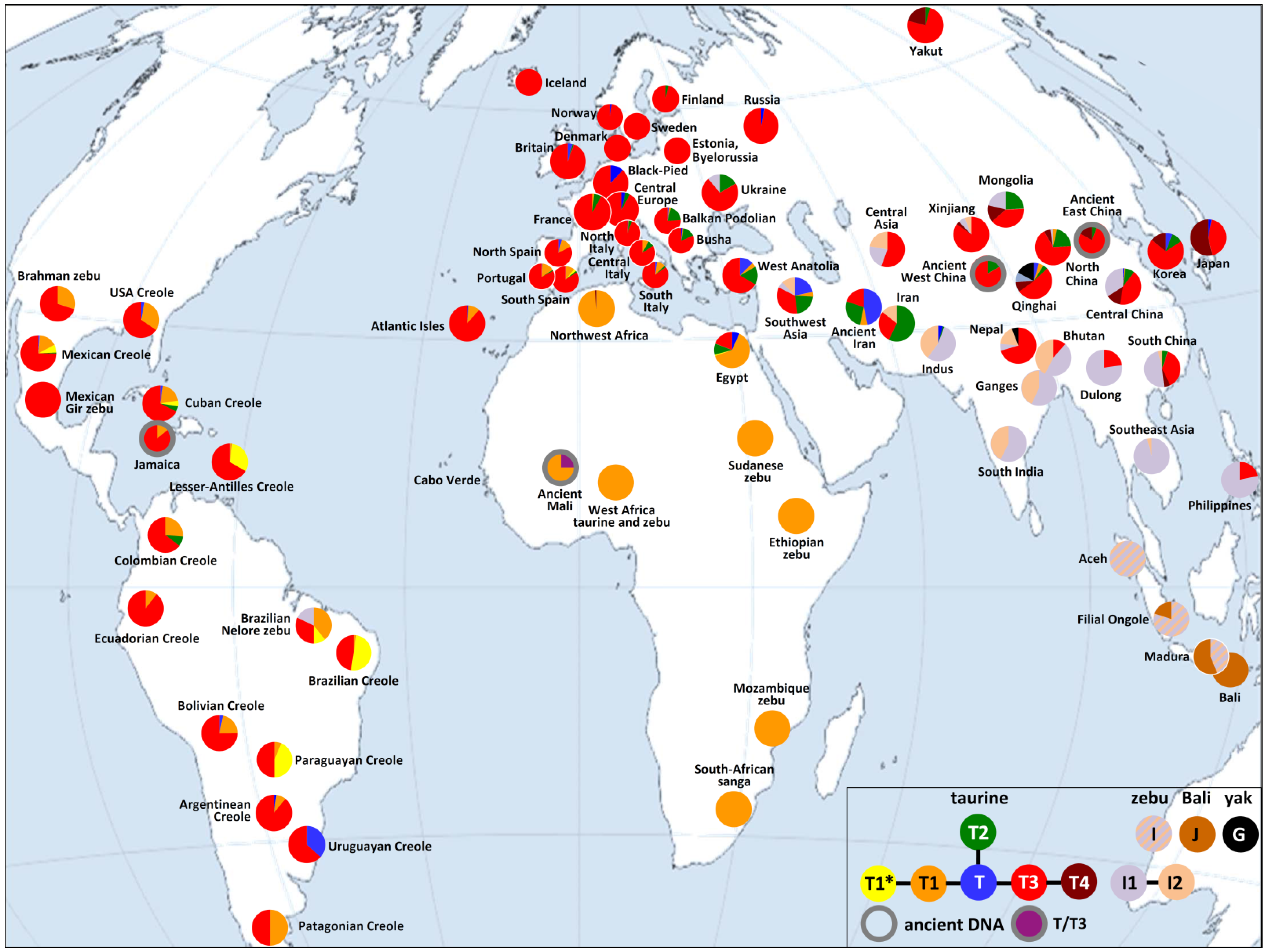
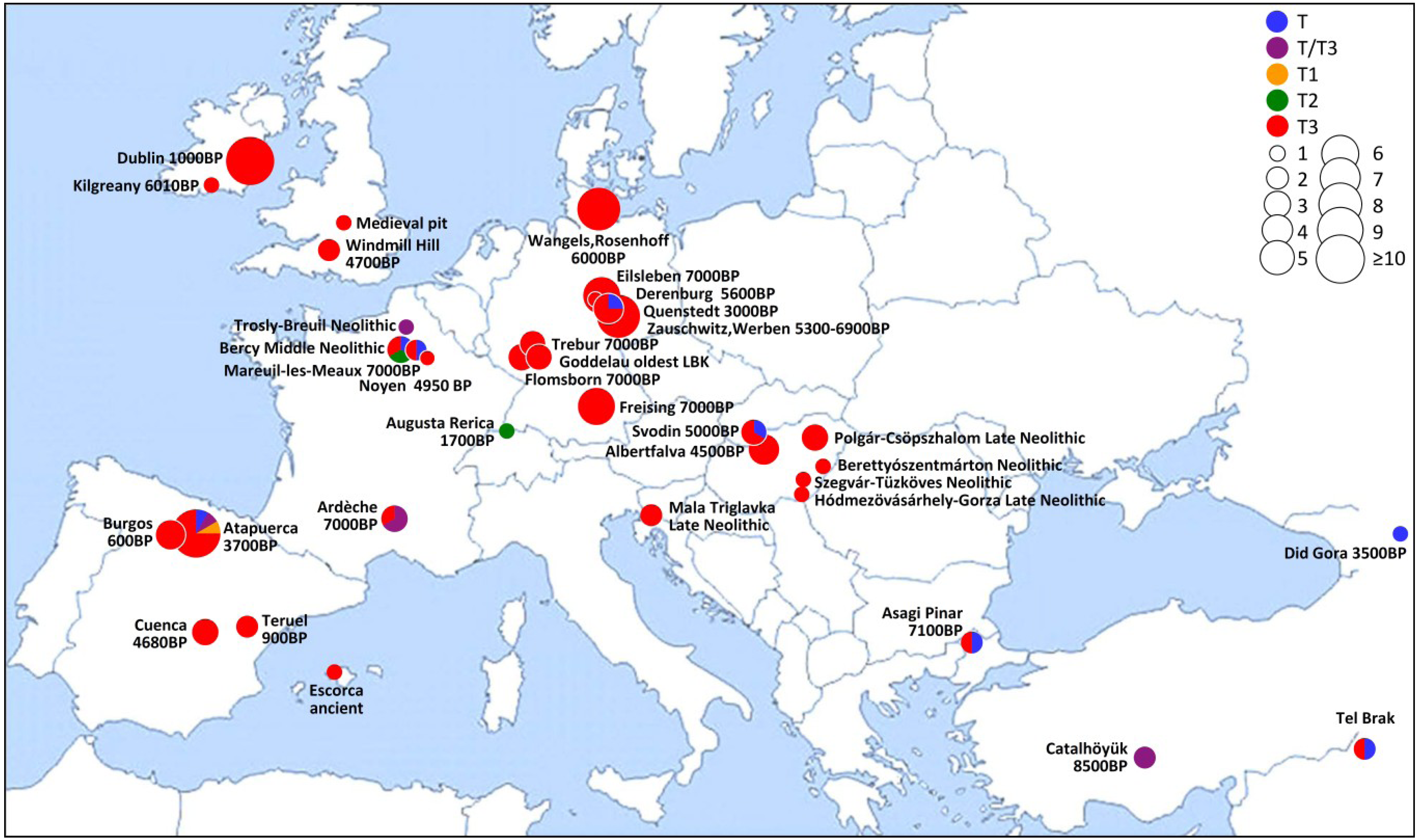
4. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of interest
References
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals: A review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef]
- Lenstra, J.A.; Groeneveld, L.F.; Eding, H.; Kantanen, J.; Williams, J.L.; Taberlet, P.; Nicolazzi, E.L.; Sölkner, J.; Simianer, H.; Ciani, E.; et al. Molecular tools and analytical approaches for the characterization of farm animal diversity. Anim. Genet. 2012, 43, 483–502. [Google Scholar] [CrossRef]
- Zeder, M.A.; Emshwiller, E.; Smith, B.D.; Bradley, D.G. Documenting domestication: The intersection of genetics and archaeology. Trends Genet. 2006, 22, 139–155. [Google Scholar] [CrossRef]
- Bradley, D.G.; Loftus, R.T.; Cunningham, C.; MacHugh, D.E. Genetics and domestic cattle origin. Evol. Anthropol. 1998, 6, 79–86. [Google Scholar] [CrossRef]
- Ajmone-Marsan, P.; Garcia, J.F.; Lenstra, J.A. On the origin of cattle: How aurochs became cattle and colonized the world. Evol. Anthropol. 2010, 19, 148–157. [Google Scholar] [CrossRef]
- Troy, C.S.; MacHugh, D.E.; Bailey, J.F.; Magee, D.A.; Loftus, R.T.; Cunningham, P.; Chamberlain, A.T.; Sykes, B.C.; Bradley, D.G. Genetic evidence for Near-Eastern origins of European cattle. Nature 2001, 410, 1088–1091. [Google Scholar] [CrossRef]
- Mannen, H.; Kohno, M.; Nagata, Y.; Tsuji, S.; Bradley, D.G.; Yeo, J.S.; Nyamsamba, D.; Zagdsuren, Y.; Yokohama, M.; Nomura, K.; et al. Independent mitochondrial origin and historical genetic differentiation in North Eastern Asian cattle. Molec. Phylogenet. Evol. 2004, 32, 539–544. [Google Scholar] [CrossRef]
- Ginja, C.; Penedo, M.C.; Melucci, L.; Quiroz, J.; Martinez Lopez, O.R.; Revidatti, M.A.; Martinez-Martinez, A.; Delgado, J.V.; Gama, L.T. Origins and genetic diversity of New World Creole cattle: Inferences from mitochondrial and Y chromosome polymorphisms. Anim. Genet. 2010, 41, 128–141. [Google Scholar] [CrossRef]
- Chen, S.; Lin, B.Z.; Baig, M.; Mitra, B.; Lopes, R.J.; Santos, A.M.; Magee, D.A.; Azevedo, M.; Tarroso, P.; Sasazaki, S.; et al. Zebu cattle are an exclusive legacy of the South Asia Neolithic. Molec. Biol. Evol. 2010, 27, 1–6. [Google Scholar]
- Achilli, A.; Olivieri, A.; Pellecchia, M.; Uboldi, C.; Colli, L.; Al-Zahery, N.; Accetturo, M.; Pala, M.; Hooshiar Kashani, B.; Perego, U.A.; et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr. Biol. 2008, 18, R157–R158. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Ginja, C.; de Gaetano, A.; Achilli, A.; Olivieri, A.; Colli, L.; Tesfaye, K.; Agha, S.H.; Gama, L.T.; Cattonaro, F.; et al. Origin and spread of Bos taurus: New clues from mitochondrial genomes belonging to haplogroup T1. PLoS One 2012, 7, e38601. [Google Scholar] [CrossRef]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusa, A.; Pala, M.; Hooshiar Kashani, B.; Perego, U.A.; Ajmone-Marsan, P.; Liotta, L.; Semino, O.; et al. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS One 2009, 4, e5753. [Google Scholar] [CrossRef]
- Stock, F.; Edwards, C.J.; Bollongino, R.; Finlay, E.K.; Burger, J.; Bradley, D.G. Cytochrome b sequences of ancient cattle and wild ox support phylogenetic complexity in the ancient and modern bovine populations. Anim. Genet. 2009, 40, 694–700. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Achilli, A.; Olivieri, A.; Negrini, R.; Colli, L.; Liotta, L.; Ajmone-Marsan, P.; Torroni, A.; Ferretti, L. The enigmatic origin of bovine mtDNA haplogroup R: Sporadic interbreeding or an independent event of Bos primigenius domestication in Italy? PLoS One 2010, 5, e15760. [Google Scholar] [CrossRef]
- Bollongino, R.; Burger, J.; Powell, A.; Mashkour, M.; Vigne, J.D.; Thomas, M.G. Modern taurine cattle descended from small number of Near-Eastern founders. Molec. Biol. Evol. 2012, 29, 2101–2104. [Google Scholar] [CrossRef]
- Bailey, J.F.; Richards, M.B.; Macaulay, V.A.; Colson, I.B.; James, I.T.; Bradley, D.G.; Hedges, R.E.; Sykes, B.C. Ancient DNA suggests a recent expansion of European cattle from a diverse wild progenitor species. Proc. Royal Soc. B: Biol. Sci. 1996, 263, 1467–1473. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Troy, C.S.; McCormick, F.; Olsaker, I.; Eythorsdottir, E.; Bradley, D.G. Early medieval cattle remains from a Scandinavian settlement in Dublin: Genetic analysis and comparison with extant breeds. Proc. Royal Soc. B: Biol. Sci. 1999, 354, 99–109. [Google Scholar]
- Edwards, C.J.; MacHugh, D.E.; Dobney, K.M.; Martin, L.; Russell, N.; Horwitz, L.K.; McIntosh, S.K.; MacDonald, K.C.; Helmer, D.; Tresset, A.; et al. Ancient DNA analysis of 101 cattle remains: Limits and prospects. J. Archaeol. Sci. 2004, 31, 695–710. [Google Scholar] [CrossRef]
- Anderung, C.; Bouwman, A.; Persson, P.; Carretero, J.M.; Ortega, A.I.; Elburg, R.; Smith, C.; Arsuaga, J.L.; Ellegren, H.; Gotherstrom, A. Prehistoric contacts over the Straits of Gibraltar indicated by genetic analysis of Iberian Bronze Age cattle. Proc. Nat. Acad. Sci. USA 2005, 102, 8431–8435. [Google Scholar] [CrossRef]
- Kühn, R.; Ludt, C.; Manhart, H.; Peters, J.; Neumair, E.; Rottmann, O. Close genetic relationship of early Neolithic cattle from Ziegelberg (Freising, Germany) with modern breeds. J. Anim. Breeding Genet. 2005, 122, 36–44. [Google Scholar] [CrossRef]
- Bollongino, R.; Edwards, C.J.; Alt, K.W.; Burger, J.; Bradley, D.G. Early history of European domestic cattle as revealed by ancient DNA. Biol. Lett. 2006, 2, 155–159. [Google Scholar] [CrossRef]
- Edwards, C.J.; Bollongino, R.; Scheu, A.; Chamberlain, A.; Tresset, A.; Vigne, J.D.; Baird, J.F.; Larson, G.; Ho, S.Y.; Heupink, T.H.; et al. Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proc. Royal Soc. B: Biol. Sci. 2007, 274, 1377–1385. [Google Scholar] [CrossRef]
- Scheu, A.; Hartz, S.; Schmölcke, U.; Tresset, A.; Burger, J.; Bollongino, R. Ancient DNA provides no evidence for independent domestication of cattle in Mesolithic Rosenhof, Northern Germany. J. Archaeol. Sci. 2008, 35, 1257–1264. [Google Scholar] [CrossRef]
- Cymbron, T.; Freeman, A.; Malheiro, M.I.; Vigne, J.D.; Bradley, D. Microsatellite diversity suggests different histories for Mediterranean and Northern European cattle populations. Proc. Royal Soc. B: Biol. Sci. 2005, 272, 1837–1843. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; Caramelli, D.; Lalueza-Fox, C.; Vernesi, C.; Ferrand, N.; Casoli, A.; Goyache, F.; Royo, L.J.; Conti, S.; Lari, M.; et al. The origin of European cattle: Evidence from modern and ancient DNA. Proc. Nat. Acad. Sci. USA 2006, 103, 8113–8118. [Google Scholar] [CrossRef]
- Pellecchia, M.; Negrini, R.; Colli, L.; Patrini, M.; Milanesi, E.; Achilli, A.; Bertorelle, G.; Cavalli-Sforza, L.L.; Piazza, A.; Torroni, A.; et al. The mystery of Etruscan origins: Novel clues from Bos taurus mitochondrial DNA. Proc. Royal Soc. B: Biol. Sci. 2007, 274, 1175–1179. [Google Scholar] [CrossRef]
- Kron, H. Roman livestock farming in southern Italy: The case against environmental determinism. In Espaces Integrés et Ressources Naturelles dans l'Empire Romain; Clavel-Lévêque, M., Hermon, E., Eds.; Presses Universitaires de Franche-Comté: Besançon, France, 2004; pp. 119–134. [Google Scholar]
- Ho, S.Y.; Larson, G.; Edwards, C.J.; Heupink, T.H.; Lakin, K.E.; Holland, P.W.; Shapiro, B. Correlating Bayesian date estimates with climatic events and domestication using a bovine case study. Biol. Lett. 2008, 4, 370–374. [Google Scholar] [CrossRef]
- Soubrier, J.; Steel, M.; Lee, M.S.; der Sarkissian, C.; Guindon, S.; Ho, S.Y.; Cooper, A. The influence of rate heterogeneity among sites on the time dependence of molecular rates. Molec. Biol. Evol. 2012, 29, 3345–3358. [Google Scholar] [CrossRef]
- Cai, D.; Sun, Y.; Tang, Z.; Hud, S.; Li, W.; Zhao, X.; Xiang, H.; Zhou, H. The origins of Chinese domestic cattle as revealed by ancient DNA analysis. J. Archaeol. Sci. 2014, 41, 423–434. [Google Scholar] [CrossRef]
- Mannen, H.; Kojima, T.; Oyama, K.; Mukai, F.; Ishida, T.; Tsuji, S. Effect of mitochondrial DNA variation on carcass traits of Japanese Black cattle. J. Anim. Sci. 1998, 76, 36–41. [Google Scholar]
- Kantanen, J.; Edwards, C.J.; Bradley, D.G.; Viinalass, H.; Thessler, S.; Ivanova, Z.; Kiselyova, T.; Cinkulov, M.; Popov, R.; Stojanovic, S.; et al. Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus). Heredity 2009, 103, 404–415. [Google Scholar] [CrossRef]
- Magee, D.A.; Meghen, C.; Harrison, S.; Troy, C.S.; Cymbron, T.; Gaillard, C.; Morrow, A.; Maillard, J.C.; Bradley, D.G. A partial African ancestry for the Creole cattle population of the Carribean. J. Hered. 2002, 93, 429–432. [Google Scholar] [CrossRef]
- Borges Lopes, M.A.; Marquez de Rezende, E.M. ABCZ História e Histórias; Câmara Brasileira do Livro: Sao Paulo, Brazil, 2001. (In Portuguese) [Google Scholar]
- Magee, D.A.; Mannen, H.; Bradley, D.G. Duality in Bos indicus mtDNA diversity: support for geographical complexity in zebu domestication. In The Evolution and History of Human Populations in South Asia; Petraglia, M.D., Allchin, B., Eds.; Springer: New York, NY, USA, 2007; pp. 385–391. [Google Scholar]
- Gou, X.; Wang, Y.; Yang, S.; Deng, W.; Mao, H. Genetic diversity and origin of Gayal and cattle in Yunnan revealed by mtDNA control region and SRY gene sequence variation. J. Anim. Breeding Genet. 2010, 127, 154–160. [Google Scholar] [CrossRef]
- Mohamad, K.; Olsson, M.; van Tol, H.T.; Mikko, S.; Vlamings, B.H.; Andersson, G.; Rodriguez-Martinez, H.; Purwantara, B.; Paling, R.W.; Colenbrander, B.; et al. On the origin of Indonesian cattle. PLoS One 2009, 4, e5490. [Google Scholar] [CrossRef]
- Takeda, K.; Satoh, M.; Neopane, S.P.; Kuwar, B.S.; Joshi, H.D.; Shrestha, N.P.; Fujise, H.; Tasai, M.; Tagami, T.; Hanada, H. Mitochondrial DNA analysis of Nepalese domestic dwarf cattle Lulu. Anim. Science J. 2003, 75, 103–110. [Google Scholar]
- Shi, J.; Qiao, H.; Hosoi, E.; Ozawa, S. Phylogenetic relationship between the Yellow cattle in Qinghai province, China, and Japanese Black cattle based on mitochondrial DNA D-loop sequence polymorphism. GenBank 2004, AB177788–AB177796. [Google Scholar]
- Yue, X.P.; Li, R.; Xie, W.M.; Xu, P.; Chang, T.C.; Liu, L.; Cheng, F.; Zhang, R.F.; Lan, X.Y.; Chen, H.; et al. Phylogeography and domestication of Chinese swamp buffalo. PLoS One 2013, 8, e56552. [Google Scholar] [CrossRef]
- Soares, P.; Abrantes, D.; Rito, T.; Thomson, N.; Radivojac, P.; Li, B.; Macaulay, V.; Samuels, D.C.; Pereira, L. Evaluating purifying selection in the mitochondrial DNA of various mammalian species. PLoS One 2013, 8, e58993. [Google Scholar] [CrossRef]
- Wang, Z.; Yonezawa, T.; Liu, B.; Ma, T.; Shen, X.; Su, J.; Guo, S.; Hasegawa, M.; Liu, J. Domestication relaxed selective constraints on the yak mitochondrial genome. Molec. Biol. Evol. 2011, 28, 1553–1556. [Google Scholar] [CrossRef]
- Paneto, J.C.; Ferraz, J.B.; Balieiro, J.C.; Bittar, J.F.; Ferreira, M.B.; Leite, M.B.; Merighe, G.K.; Meirelles, F.V. Bos indicus or Bos taurus mitochondrial DNA—Comparison of productive and reproductive breeding values in a Guzerat dairy herd. Genet. Molec. Res. 2008, 7, 592–602. [Google Scholar] [CrossRef]
- Horsburgh, K.A.; Prost, S.; Gosling, A.; Stanton, J.A.; Rand, C.; Matisoo-Smith, E.A. The genetic diversity of the Nguni breed of African cattle (Bos spp.): Complete mitochondrial genomes of haplogroup T1. PLoS One 2013, 8, e71956. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lenstra, J.A.; Ajmone-Marsan, P.; Beja-Pereira, A.; Bollongino, R.; Bradley, D.G.; Colli, L.; De Gaetano, A.; Edwards, C.J.; Felius, M.; Ferretti, L.; et al. Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle. Diversity 2014, 6, 178-187. https://doi.org/10.3390/d6010178
Lenstra JA, Ajmone-Marsan P, Beja-Pereira A, Bollongino R, Bradley DG, Colli L, De Gaetano A, Edwards CJ, Felius M, Ferretti L, et al. Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle. Diversity. 2014; 6(1):178-187. https://doi.org/10.3390/d6010178
Chicago/Turabian StyleLenstra, Johannes A., Paolo Ajmone-Marsan, Albano Beja-Pereira, Ruth Bollongino, Daniel G. Bradley, Licia Colli, Anna De Gaetano, Ceiridwen J. Edwards, Marleen Felius, Luca Ferretti, and et al. 2014. "Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle" Diversity 6, no. 1: 178-187. https://doi.org/10.3390/d6010178
APA StyleLenstra, J. A., Ajmone-Marsan, P., Beja-Pereira, A., Bollongino, R., Bradley, D. G., Colli, L., De Gaetano, A., Edwards, C. J., Felius, M., Ferretti, L., Ginja, C., Hristov, P., Kantanen, J., Lirón, J. P., Magee, D. A., Negrini, R., & Radoslavov, G. A. (2014). Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle. Diversity, 6(1), 178-187. https://doi.org/10.3390/d6010178







