Molecular Identification and Historic Demography of the Marine Tucuxi (Sotalia guianensis) at the Amazon River’s Mouth by Means of Mitochondrial Control Region Gene Sequences and Implications for Conservation
Abstract
:1. Introduction
2. Material and Methods
2.1. Samples and Molecular Procedures
2.2. Data Analysis
2.2.1. Gene Diversity
2.2.2. Demographic Changes
2.2.3. Phylogenetic Analyses and Temporal Split Times
3. Results
3.1. Gene Diversity in the Population of S. guianensis at the Mouth of the Amazon River
3.2. Possible Demographic Changes in the History of the S. guianensis Population within the Amazon River Estuary
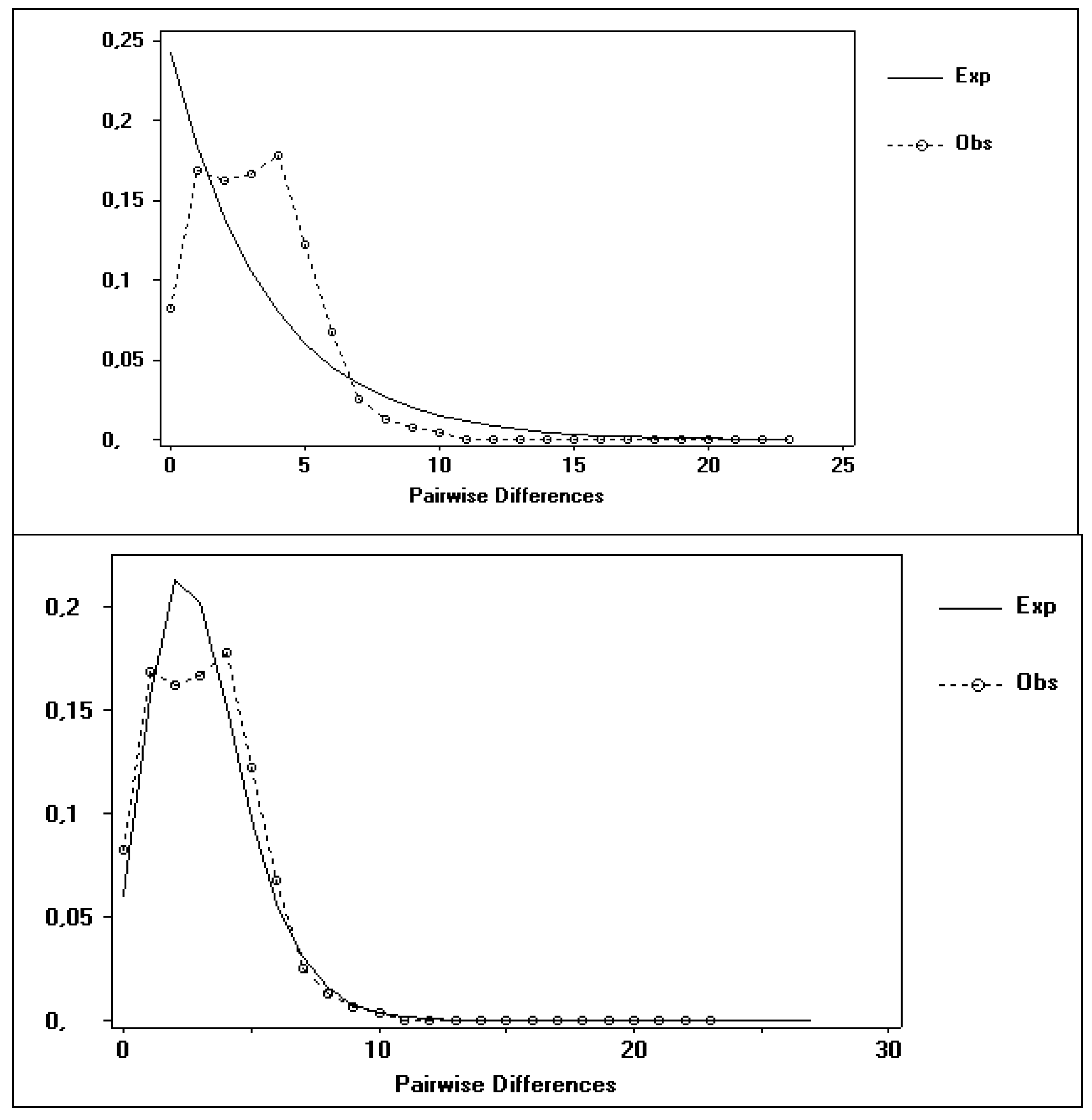


3.3. Phylogenetic Analyses and Temporal Divergence Splits among the mt Haplotype Lineages Found in the S. guianensis Population Studied in the Amazon River Mouth
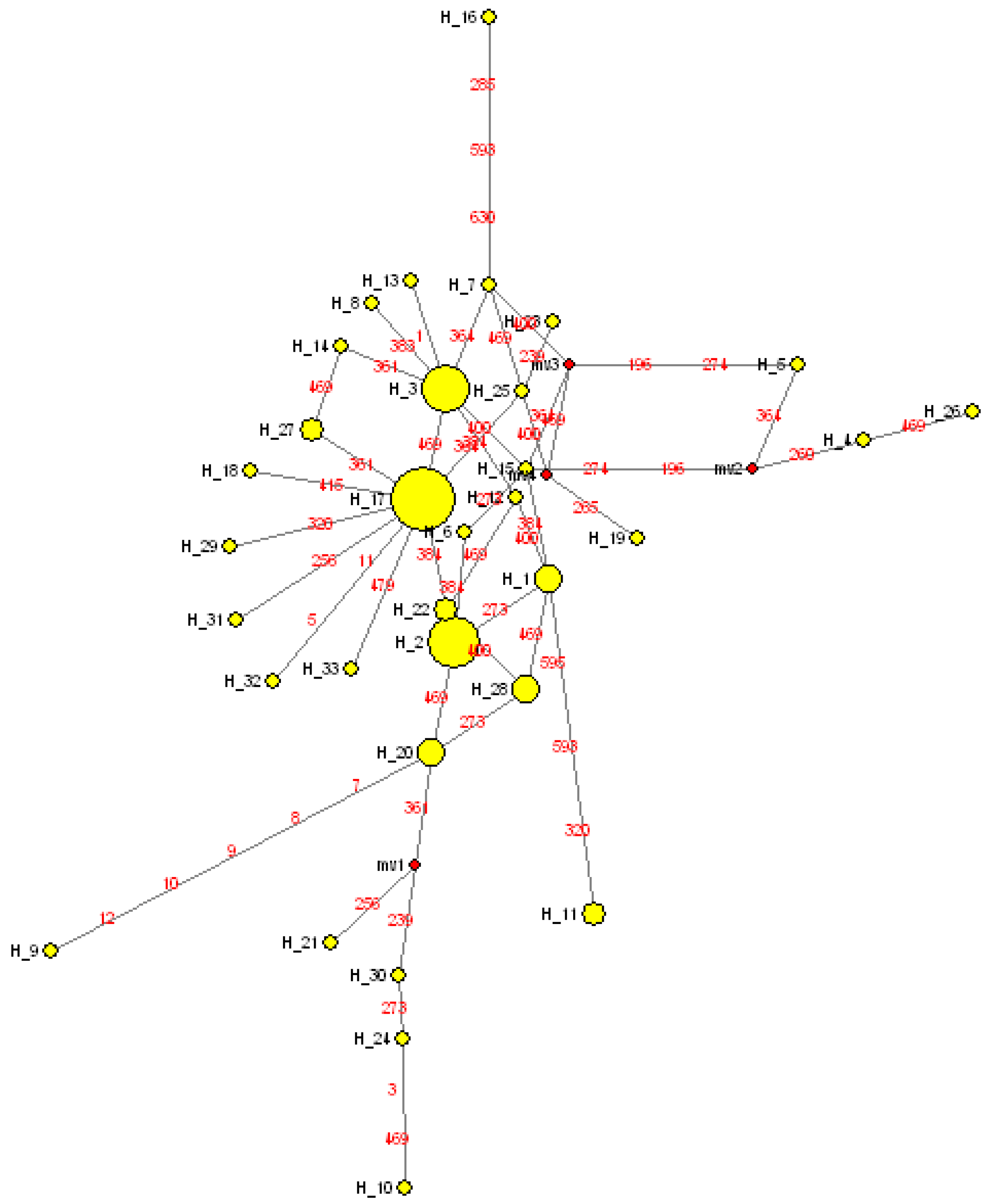
4. Discussion
4.1. Gene Diversity
4.2. Historical Demographic Evolution of the S. guianensis Population at the Amazon River Mouth and Proliferation of Haplotype Lineages
Acknowledgments
Conflicts of Interest
References
- Borobia, M.; Siciliano, S.; Lodi, L.; Whoek, I. Distribution of the South American dolphin Sotalia fluviatilis. Can. J. Zool. 1991, 69, 1025–1039. [Google Scholar] [CrossRef]
- Carr, T.; Bonde, R. Tucuxi (Sotalia fluviatilis) occurs in Nicaragua, 800 km north of its previously known range. Mar. Mamm. Sci. 2000, 16, 447–452. [Google Scholar] [CrossRef]
- Da Silva, V.M.F.; Best, R.C. Sotalia fluviatilis. Mamm. Spec. 1996, 527, 1–7. [Google Scholar] [CrossRef]
- Robineau, D. Les types de cétacés actuels du Muséum National D´Histoire Naturelle II. Delphinidae, Phocoenidae. Bulletin du Muséum National d’Histoire Naturelle Paris 1990, 12, 197–238. [Google Scholar]
- Hershkovitz, P. Catalog of living whales. Bulletin of the United States National Museum; Smithsonian Institution: Washington, DC, 1966; Volume 246, pp. 1–259. [Google Scholar]
- Flower, W.H. On the characters and divisions of the family Delphinidae. Proc. Zool. Soc. Lond. 1883, 43, 466–513. [Google Scholar]
- Hershkovitz, P. Notes on South American dolphins of the genera Inia, Sotalia and Tursiops. J. Mamm. 1962, 44, 98–103. [Google Scholar] [CrossRef]
- Cabrera, A. Catálogo de los mamíferos de América del Sur, II, (Sirenia-Perissodactyla-Artiodactyla-Lagomorpha-Cetacea). Revista del Museo Argentino de Ciencias Naturales “Bernardino Rivadavia.” Ciencias Zool. 1961, 4, 309–732. [Google Scholar]
- Mitchell, E.D. Report of the meeting on smaller cetaceans. J. Fish Res. Board Can. 1975, 32, 889–983. [Google Scholar] [CrossRef]
- Leatherwood, S.; Reeves, R.R. The Sierra Club Handbook of Whales and Dolphins; Sierra Club Books: San Francisco, CA, USA, 1983. [Google Scholar]
- Borobia, M. Distribution and Morphometrics of South American Dolphins of the Genus Sotalia. Master’s Thesis, McGill University, Montreal, MN, Canada, 7 March 1989; p. 81. [Google Scholar]
- Rice, D.W. Marine Mammals of the World: Systematics and Distribution; Special Publication Number 4; The Society of Marine Mammalogy: Lawrence, KS, USA, 1998. [Google Scholar]
- Flores, P.A.C. Tucuxi—Sotalia fluviatilis. In Encyclopedia of Marine Mammals; Perrin, W.F., Würsig, B., Thewissen, J.G.M., Eds.; Academic: San Diego, FL, USA, 2002; pp. 1267–1269. [Google Scholar]
- Furtado-Neto, M.A.A. Molecular systematics and population genetics of marine vertebrates from Brazil. Ph.D. Thesis., Memorial University of Newfoundland, Newfoundland, Canada, 1998. [Google Scholar]
- Monteiro-Filho, E.L.D.A.; Rabello-Monteiro, L.; Reis, S.F.D. Skull shape and size divergence in dolphins of the genus Sotalia: A morphometric tridimensional analysis. J. Mammal. 2002, 83, 125–134. [Google Scholar] [CrossRef]
- Cunha, H.A.; Da Silva, V.M.F.; Lailson-Brito, J.J.; Santos, M.C.O.; Flores, P.A.C.; Martin, A.R.; Azevedo, A.F.; Fragoso, A.B.L.; Zanelatto, R.C.; Solé-Cava, A.M. Riverine and marine ecotypes of Sotalia fluviatilis are different species. Mar. Biol. 2005, 148, 449–457. [Google Scholar] [CrossRef]
- Caballero, S.; Trujillo, F.; Vianna, J.A.; Barrios-Garrido, H.; Montiel, M.G.; Beltrán-Pedreros, S.; Marmontel, M.; Santos, M.C.; Rossi-Santos, M.; Santos, F.R.; et al. Taxonomic status of the genus Sotalia: Species level ranking for “tucuxi” (Sotalia fluviatilis) and “costero” (Sotalia guianensis) dolphins. Mar. Mamm. Sci. 2007, 23, 358–386. [Google Scholar] [CrossRef]
- Boher, S.; Bolaños, J.; Cova, L.J. Sobre un avistamiento del delfín estuarino o bufete (Sotalia fluviatilis) en el Orinoco Medio. Acta Científica Venezolana 1995, 46, 217–218. [Google Scholar]
- Cunha, H.C.; Da Silva, V.M.F.; Solé-Cava, A.M. Molecular Ecology and Systematics of Sotalia Dolphins. In Biology, Evolution and Conservation of River Dolphins within South America and Asia; Ruiz-Garcia, M., Shostell, J., Eds.; Nova Science: New York, NY, USA, 2010; pp. 261–283. [Google Scholar]
- Cunha, H.A. Sistemática molecular e filogeografia do gênero Sotalia Gray 1866 (Delphinidae) no Brasil. Ph.D. Thesis, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil, 17 June 2007. [Google Scholar]
- Cunha, H.A.; Solé-Cava, A.M. Phylogeography of Sotalia guianensis along the Brazilian coast. In Sociedad Latinoamericana de Especialistas en Mamíferos Acuáticos – SOLAMAC; (in Portuguese). Workshop Internacional sobre Pesquisa e Conservação dos Golfinhos do Gênero Sotalia: Búzios, Brazil, 2006. [Google Scholar]
- Hollatz, C.; Flach, L.; Scott Baker, C.; Santos, F.R. Microsatellite data reveal fine genetic structure in male Guiana dolphins (Sotalia guianensis) in two geographically close embayments at south-eastern coast of Brazil. Mar. Biol. 2011, 158, 927–933. [Google Scholar] [CrossRef]
- Caballero, S. Genetic Characterization of the South American Coastal and Riverine Dolphins Sotalia. Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 2006; p. 187. [Google Scholar]
- Sambrock, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1989. [Google Scholar]
- Shields, G.F.; Kocher, T.D. Phylogenetic relationships of North American Ursids based on analysis of mitochondrial DNA. Evolution 1991, 45, 218–221. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmouguin, F.; Higgins, D.G. The Clustal-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 1997, 24, 4876–4882. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H.C. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Rogers, A.R.; Fraley, A.E.; Bamshad, M.J.; Watkins, W.S.; Jorde, L.B. Mitochondrial mismatch analysis is insensitive to the mutational process. Mol. Biol. Evol. 1996, 13, 895–902. [Google Scholar] [CrossRef]
- Harpending, H.C.; Sherry, S.T.; Rogers, A.R.; Stoneking, M. Genetic structure of ancient human populations. Curr. Antrophol. 1993, 34, 483–496. [Google Scholar]
- Harpending, H. Signature of ancient population growth in a low resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar]
- Fu, Y.X. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar]
- Simonsen, K.L.; Churchill, G.A.; Aquadro, C.F. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 1995, 141, 413–429. [Google Scholar]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer v1.4. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 2 October 2012).
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Cunha, H.C.; Moraes, L.C.; Medeiros, B.V.; Lailson-Brito, J., Jr.; Da Silva, V.M.F.; Solé-Cava, A.M.; Schrago, C.G. Phylogenetic status and timescale for the diversification of Steno and Sotalia dolphins. PLoS One 2011, 6, e28297. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic analysis using parsimony and other methods. pp. 1–142. Available online: http://www.paup.csit.fsu.edu (accessed on 7 January 2002).
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Morral, N.; Bertranpetit, J.; Estivill, X. The origin of the major cystic fibrosis mutation (delta F508) in European populations. Nat. Genet. 1994, 7, 169–175. [Google Scholar] [CrossRef]
- Saillard, J.; Forster, P.; Lynnerup, N.; Bandelt, H.-J.; Norby, S. mtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am. J. Hum. Genet. 2000, 67, 718–726. [Google Scholar] [CrossRef]
- Beltran-Pedreros, S.; Petrere, M.; Filgueiras-Henriques, L.A. Ethnoecology of Sotalia guianensis (Gervais, 1853) in the Amazon Estuary. In Biology, Evolution and Conservation of River Dolphins within South America and Asia; Ruiz-Garcia, M., Shostell, J., Eds.; Nova Science: New York, NY, USA, 2010; pp. 221–235. [Google Scholar]
- Muller-Karger, F.E.; Mcclain, C.R.; Richardson, P.L. The dispersal of the Amazon’s water. Nature 1988, 333, 57–59. [Google Scholar]
- Gravena, W.; Hrbek, T.; Da Silva, V.M.F.; Farias, I.P. Amazon River dolphin love fetishes: From folklore to molecular forensics. Mar. Mamm. Sci. 2008, 24, 969–978. [Google Scholar]
- Sholl, T.G.C.; Nascimento, F.F.; Leoncini, O.; Bonvicino, C.R.; Siciliano, S. Taxonomic identification of dolphin love charms commercialized in the Amazonian region through the analysis of cytochrome b DNA. J. Mar. Biol. Assoc. UK 2008, 88, 1207–1210. [Google Scholar]
- Pinedo, M.C. Intentional and incidental capture of marine mammals in fishing nets. In Conclusiones de la primera reunión de trabajo de expertos en mamíferos acuáticos de América del Sur; Fundación Vida Silvestre Argentina Editors: Buenos Aires, Argentina, 1985; pp. 1–236. [Google Scholar]
- Flach, L.; Flach, P.A.; Chiarello, A.G. Aspects of behavioral ecology of Sotalia guianensis in Sepetiba Bay, southeast Brazil. Mar. Mamm. Sci. 2008, 24, 503–515. [Google Scholar] [CrossRef]
- Hoelzel, A.R.; Goldsworthy, S.D.; Fleischer, R.C. Population genetics. In Marine Mammal Biology: An Evolutionary Approach; Hoelzel, R., Ed.; Blackwell Science: Oxford, England, 2002; pp. 325–352. [Google Scholar]
- Caballero, S.; Trujillo, F.; Ruiz-García, M.; Vianna, J.; Marmontel, M.; Santos, F.R.; Baker, C.S. Population structure and phylogeography of tucuxi dolphins (Sotalia fluviatilis). In Biology, Evolution, and Conservation of River Dolphins within South America and Asia; Ruiz-García, M., Shostell, J., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2010; pp. 285–299. [Google Scholar]
- Borobia, M.; Rosas, F.C.W. Tucuxi, Sotalia fluviatilis. In Estado de Conservación de los Mamíferos Marinos del Atlántico Sudoccidental; (in Spanish). Informes y estudios del Programa de Mares Regionales del PNUMA No 138; Capozzo, H.L., Junìn, M., Eds.; SSC Editorial: Nairobi, Kenya, 1991; pp. 36–41. [Google Scholar]
- Magnusson, W.E.; Best, R.C.; Da Silva, V.M.F. Numbers and behaviour of Amazonian dolphins, Inia geoffrensis and Sotalia fluviatilis in the Rio Solimoes, Brasil. Aquat. Mamm. 1988, 8, 27–41. [Google Scholar]
- Vidal, O.; Barlow, J.; Hurtado, L.A.; Torre, J.; Cendón, P.; Ojeda, Z. Distribution and abundance of the Amazon river dolphin (Inia geoffrensis) and the Tucuxi (Sotalia fluviatilis) in the upper Amazon river. Mar. Mamm. Sci. 1997, 13, 427–445. [Google Scholar] [CrossRef]
- Hoelzel, A.R.; Hancock, J.M.; Dover, G.A. Evolution of the cetacean mitochondrial D-loop region. Mol. Biol. Evol. 1991, 8, 475–493. [Google Scholar]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef]
- Hoorn, C.J.; Guerrero, G.A.; Sarmiento, D.; Lorente, M.A. Andean tectonics as a cause for changing drainage patterns in miocene Northern South-America. Geology 1995, 23, 237–240. [Google Scholar] [CrossRef]
- Lundberg, J.G.; Marshall, L.G.; Guerrero, J.; Horton, B.; Malabarba, M.C.S.L.; Wesseling, F. The Stage for Neotropical Fish Diversification: A History of Tropical South American Rivers. In Phylogeny and Classification of Neotropical Fishes; Malabarba, L.R., Reis, R.E., Vari, R.P., Lucena, Z.M., Lucena, C.A.S., Eds.; Edipucrs: Porto Alegre, Brasil, 1998; p. 603. [Google Scholar]
- Klammer, G. The Relief of the Extra-Andean Amazon Basin. In The Amazon: Limnology and Landscape Ecology of a Mighty Tropical River and its Basin; Sioli, H., Ed.; Dr W Junk Publisher: Dordrecht, The Netherlands, 1984; pp. 47–83. [Google Scholar]
- McGowen, M.R.; Spaulding, M.; Gatesy, J. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 2009, 53, 891–906. [Google Scholar] [CrossRef]
- Steeman, M.E.; Hebsgaard, M.B.; Fordyce, R.E.; Ho, S.Y.W.; Rabosky, D.L.; Nielsen, R.; Rahbek, C.; Glenner, H.; Sørensen, M.V.; Willerslev, E. Radiation of extant cetaceans driven by restructuring of the oceans. Syst. Biol. 2009, 58, 573–585. [Google Scholar] [CrossRef]
- Forasiepi, A.; Martinelli, A.; Blanco, J. Bestiario Fósil. Mamíferos del Pleistoceno de la Argentina; Editorial Albatros: Buenos Aires, Argentina, 2007; pp. 1–190. [Google Scholar]
- Van Der Hammen, T. La Paleoecología de Suramérica Tropical. Cuarenta años de Investigación de la Historia del Medio Ambiente y de la Vegetación. In Historia, Ecología y Vegetación; Corporación Colombiana para la Amazonía-Araracuara: Bogotá, Colombia, 1992; pp. 1–411. [Google Scholar]
- Campbell, K.E.; Frailey, C.D.; Romero-Pittman, L. The Pan-Amazonian Ucayali Peneplain, late Neogene sedimentation in Amazonia, and the birth of the modern Amazon River system. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2006, 239, 166–219. [Google Scholar] [CrossRef]
- Figueiredo, J.; Hoorn, C.; van Der Ven, P.; Soares, E. Late Miocene onset of the Amazon River and the Amazon deep-sea fan: Evidence from the Foz do Amazonas Basin. Geology 2009, 37, 619–622. [Google Scholar] [CrossRef]
- Haq, B.U.; Hardenbol, J.; Vail, P.R. Chronology of fluctuating sea levels since the Triassic. Science 1987, 235, 1156–1167. [Google Scholar]
- Díaz De Gamero, M.L. The changing course of the Orinoco River during the Neogene: A review. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1996, 123, 385–402. [Google Scholar] [CrossRef]
- Cossíos, E.D.; Lucherini, M.; Ruiz-García, M.; Angers, B. Influence of ancient glacial periods on the Andean fauna: The case of the Pampas cat (Leopardus colocolo). BMC Evol. Biol. 2009, 9, 68–79. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Pinedo-Castro, M. Population Genetics and Phylogeographic Analyses of the Jaguarundi (Puma yagouaroundi) by Means of Three Mitochondrial Markers: The First Molecular Population Study of This Species. In Molecular Population Genetics, Phylogenetics, Evolutionary Biology and Conservation of the Neotropical Carnivores; Ruiz-Garcia, M., Shostell, J.M., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 245–288. [Google Scholar]
- Ruiz-García, M.; Rivas-Sánchez, D.; Lichilín-Ortiz, N. Phylogenetics Relationships among Four Putative Taxa of Foxes of the Pseudoalopex Genus (Canidae, Carnivora) and Molecular Population Genetics of Ps. culpaeus and Ps. sechurae . In Molecular Population Genetics, Phylogenetics, Evolutionary Biology and Conservation of the Neotropical Carnivores; Ruiz-García, M., Shostell, J., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 97–128. [Google Scholar]
- Absy, M.L.; Cleff, A.M.; Fournier, M.; Martin, L.; Servant, M.; Sifeine, A.; da Silva, M.F.; Soubies, F.; Suguio, K.; Turc, B.; et al. Mise en evidence de quatre phases d’overture de la fôret dense dans le Sud-Est de l’Amazone en cours des 60,000 dernières années. Compte Rendeu Academie de Sciences, Serie 2, Paris 1991, 312, 673–678. [Google Scholar]
- Van Der Hammen, T.; Absy, M.L. Amazonia during the last Glacial. Palaeogegr. Palaeoclimatol. Palaeoecol. 1994, 109, 247–261. [Google Scholar] [CrossRef]
- Liu, K.; Colinvaux, A. Forest changes in the Amazon basin during the last glacial maximum. Nature 1985, 318, 556–557. [Google Scholar] [CrossRef]
- Colinvaux, P.A.; Liu, K. The Late Quaternary Climate of the Western Amazon Basin. In Abrupt Climatic Change; Berger, W., Labegrie, L., Eds.; Riedel: Dordrech, the Netherlands, 1987; pp. 113–122. [Google Scholar]
- MaCneish, R.S. The Early Man Remains from Pikimachay Cave, Ayacucho Basin, Highland Peru. In Pre-Llano cultures of the Americas: Paradoxes and Possibilities; Humprey, R.L., Standford, D., Eds.; Anthropological Society of Washington: Washington, DC, USA, 1979; pp. 1–47. [Google Scholar]
- Pereira, S.D.; Chaves, H.A.F.; Coelho, L.G. The little ice age in Sepetiba Bay, Rio de Janeiro–Brazil. J. Coast. Res. 2009, 56, 252–256. [Google Scholar]
- Krützen, M.; Sherwin, W.B.; Berggren, P.; Gales, N. Population structure in an inshore cetacean revealed by microsatellite and mtDNA analysis: Bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Mar. Mamm. Sci. 2004, 20, 28–47. [Google Scholar] [CrossRef]
- Natoli, A.; Birkun, A.; Aguilar, A.; Lopez, A.; Hoelzel, A.R. Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc. R. Soc. Lond. B 2005, 272, 1217–1226. [Google Scholar] [CrossRef]
- Möller, L.M.; Wiszniewski, J.; Allen, S.J.; Beheregaray, L.B. Habitat type promotes rapid and extremely localized genetic differentiation in dolphins. Mar. Freshwater Res. 2007, 58, 640–648. [Google Scholar] [CrossRef]
- Wiszniewsky, J.; Beheregaray, L.B.; Allen, S.J.; Möller, L.M. Environmental and social influences on the genetic structure of bottlenose dolphins (Tursiops aduncus) in South-eastern Australia. Conserv. Genet. 2010, 11, 1405–1419. [Google Scholar] [CrossRef]
- Waples, R.S. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 1989, 121, 379–391. [Google Scholar]
- Ruiz-García, M. Genética de Poblaciones: Teoría y aplicación a la conservación de mamíferos neotropicales (Oso andino y delfín rosado). Boletín de la Real Sociedad Española de Historia Natural 2007, 102, 99–126. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ruiz-García, M.; Mejia, D.; Escobar-Armel, P.; Tejada-Martínez, D.; Shostell, J.M. Molecular Identification and Historic Demography of the Marine Tucuxi (Sotalia guianensis) at the Amazon River’s Mouth by Means of Mitochondrial Control Region Gene Sequences and Implications for Conservation. Diversity 2013, 5, 703-723. https://doi.org/10.3390/d5040703
Ruiz-García M, Mejia D, Escobar-Armel P, Tejada-Martínez D, Shostell JM. Molecular Identification and Historic Demography of the Marine Tucuxi (Sotalia guianensis) at the Amazon River’s Mouth by Means of Mitochondrial Control Region Gene Sequences and Implications for Conservation. Diversity. 2013; 5(4):703-723. https://doi.org/10.3390/d5040703
Chicago/Turabian StyleRuiz-García, Manuel, David Mejia, Pablo Escobar-Armel, Daniela Tejada-Martínez, and Joseph Mark Shostell. 2013. "Molecular Identification and Historic Demography of the Marine Tucuxi (Sotalia guianensis) at the Amazon River’s Mouth by Means of Mitochondrial Control Region Gene Sequences and Implications for Conservation" Diversity 5, no. 4: 703-723. https://doi.org/10.3390/d5040703
APA StyleRuiz-García, M., Mejia, D., Escobar-Armel, P., Tejada-Martínez, D., & Shostell, J. M. (2013). Molecular Identification and Historic Demography of the Marine Tucuxi (Sotalia guianensis) at the Amazon River’s Mouth by Means of Mitochondrial Control Region Gene Sequences and Implications for Conservation. Diversity, 5(4), 703-723. https://doi.org/10.3390/d5040703



