Functional Analysis of the Drosophila Dnop5 Using Targeted RNA Interference
Abstract
:1. Introduction
2. Results and Discussion
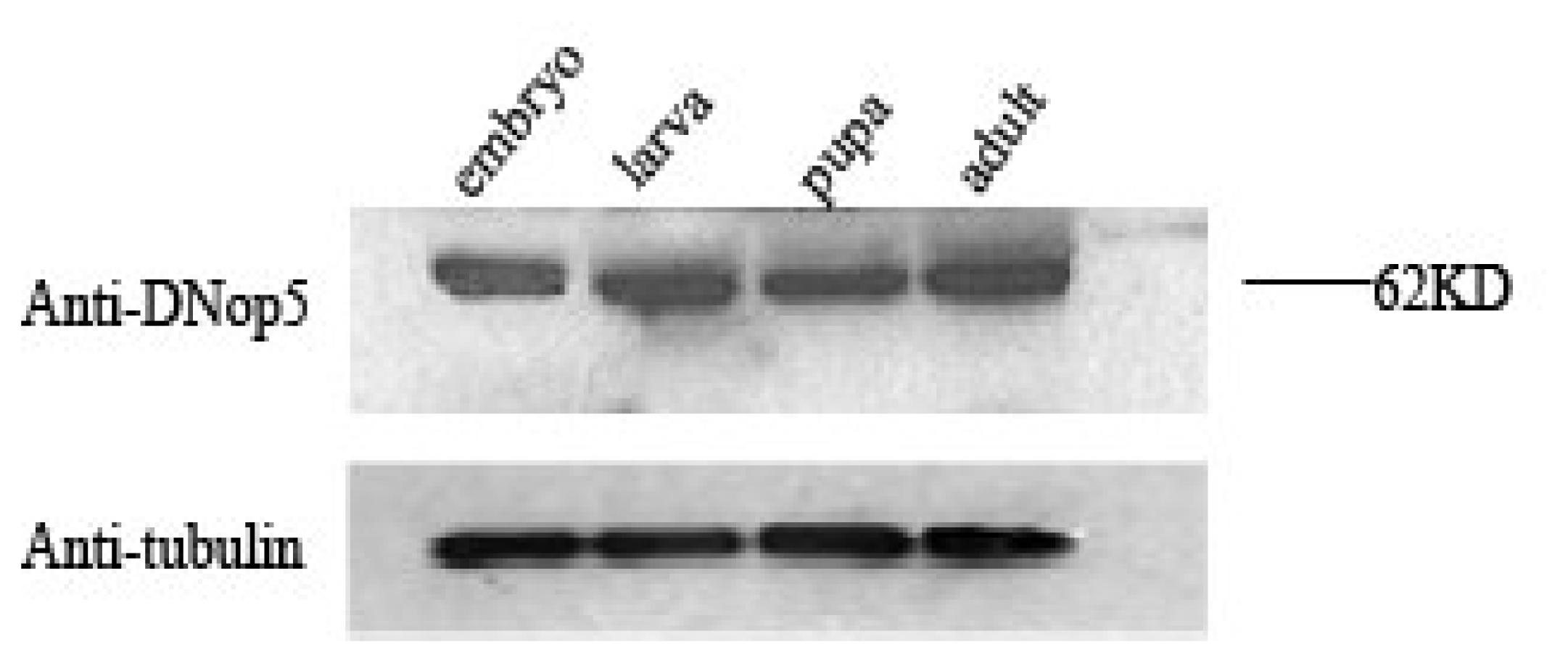
2.1. DNop5 is ubiquitously expressed throughout Drosophila development
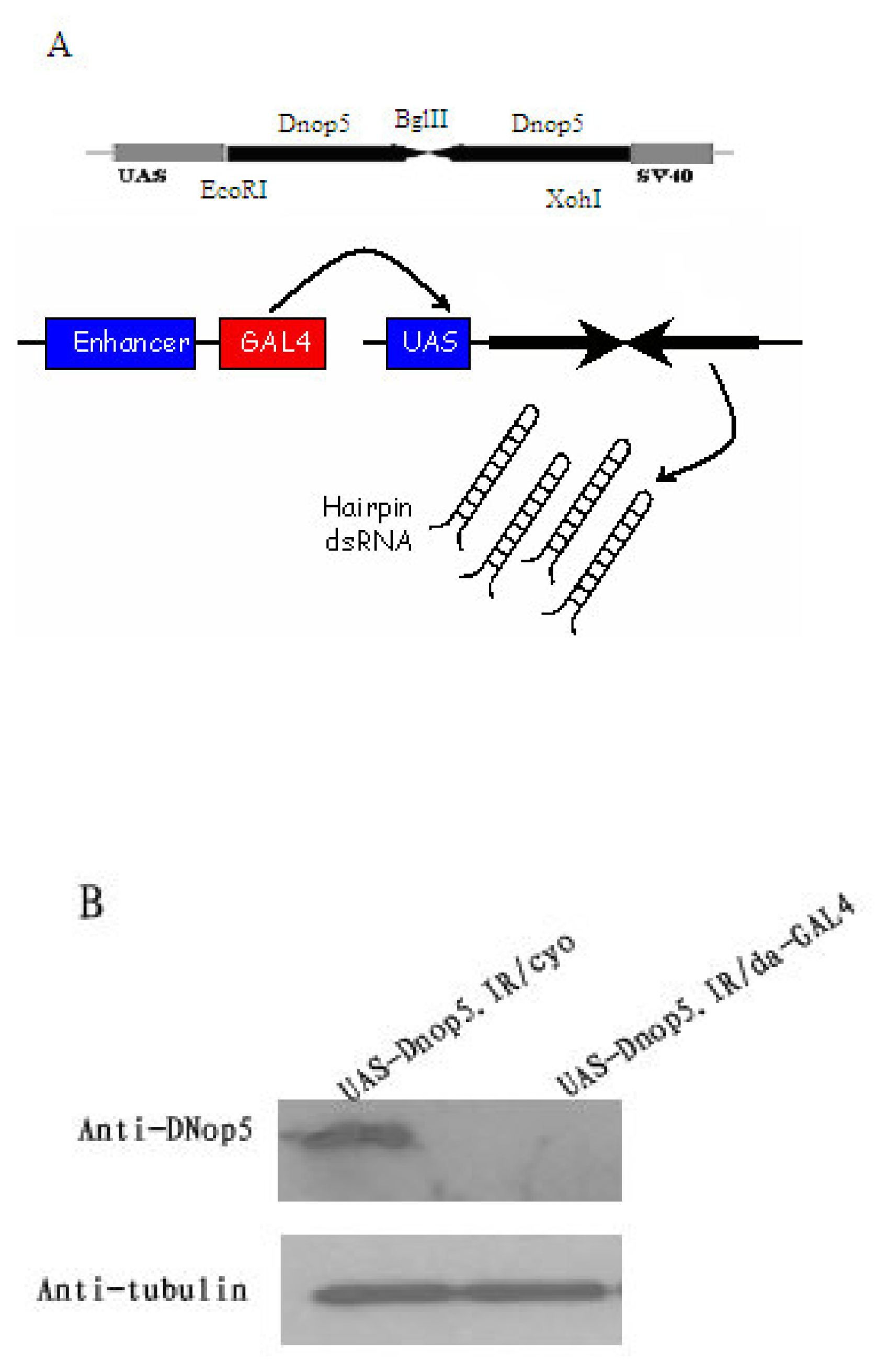
2.2 Inducible RNAi targets Dnop5
2.3 Phenotypic analysis of Dnop5 RNAi mutant flies
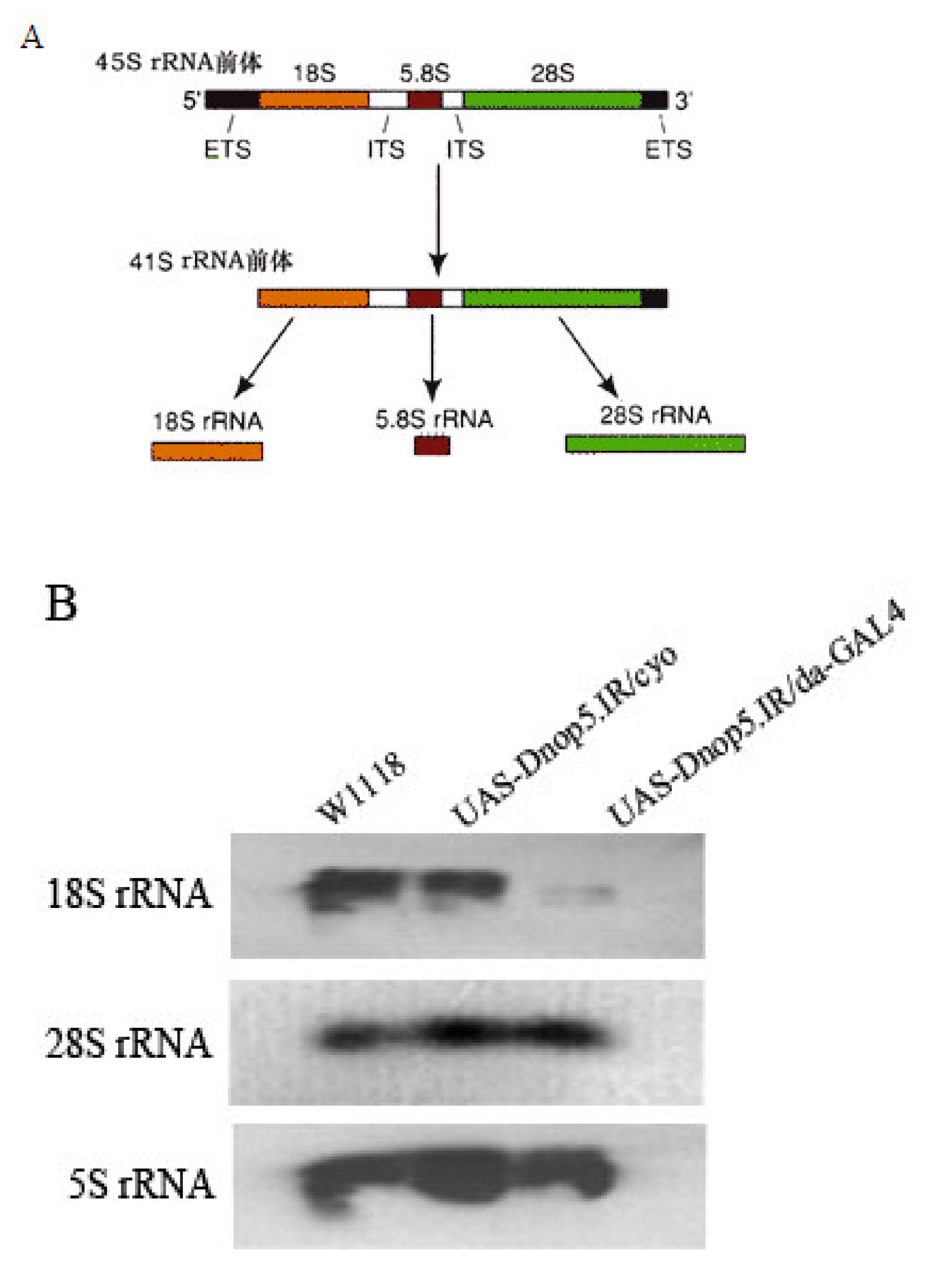
2.4 Depletion of Dnop5 impairs synthesis of 18S rRNA
3. Experimental Section
3.1 Fly stocks
3.2 Plasmid construction
3.3 Transgenic Fly Lines
3.4 Antibody generation
3.5 Western blot analysis
3.6 RNA isolation and northern blot analysis

Acknowledgements
References and Notes
- Gonzales, F.A.; Zanchin, N.I.; Luz, J.S.; Oliveira, C.C. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in pre-rRNA processing. J. Mol. Biol 2005, 346, 437–455. [Google Scholar]
- Lyman, S.K.; Gerace, L.; Baserga, S.J. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 1999, 5, 1597–1604. [Google Scholar]
- Wu, P.; Brockenbrough, J.S.; Metcalfe, A.C.; Chen, S.; Aris, J.P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem 1998, 273, 16453–16463. [Google Scholar]
- Vorbruggen, G.; Onell, S.; Jackle, H. Restricted expression and subnuclear localization of the Drosophila gene Dnop5, a member of the Nop/Sik family of the conserved rRNA processing factors. Mech. Dev 2000, 90, 305–308. [Google Scholar]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar]
- Lambertsson, A. The minute genes in Drosophila and their molecular functions. Adv. Genet 1998, 38, 69–134. [Google Scholar]
- Cramton, S.E.; Laski, F.A. String of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 1994, 137, 1039–1048. [Google Scholar]
- Schmidt, A.; Hollmann, M.; Schafer, U. A newly identified Minute locus, M(2)32D, encodes the ribosomal protein L9 in Drosophila melanogaster. Mol. Gen. Genet 1996, 251, 381–387. [Google Scholar]
- Enerly, E.; Larsson, J.; Lambertsson, A. Reverse genetics in Drosophila: from sequence to phenotype using UAS-RNAi transgenic flies. Genesis 2002, 34, 152–155. [Google Scholar]
- Enerly, E.; Larsson, J.; Lambertsson, A. Silencing the Drosophila ribosomal protein L14 gene using targeted RNA interference causes distinct somatic anomalies. Gene 2003, 320, 41–48. [Google Scholar]
- Van Roessel, P.; Hayward, N.M.; Barros, C.S.; Brand, A.H. Two-color GFP imaging demonstrates cell-autonomy of GAL4-driven RNA interference in Drosophila. Genetics 2002, 160, 637–648. [Google Scholar]
- Kennerdell, J.R.; Carthew, R.W. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol 2000, 18, 896–898. [Google Scholar]
- Nishinaka, N.; Hongo, S.; Zhou, C.J.; Shioda, S.; Takahashi, R.; Yamauchi, Y.; Ohashi, T.; Ohki, T.; Nakada, N.; Takeda, F.; Takeda, M. Identification of the novel developmentally regulated gene, Bdm2, which is highly expressed in fetal rat brain. Devel. Brain Res 2000, 120, 57–64. [Google Scholar]
© 2007 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Zhang, Y.; Ding, J.; Wan, Y.; Xie, W.; Yuan, L. Functional Analysis of the Drosophila Dnop5 Using Targeted RNA Interference. Int. J. Mol. Sci. 2007, 8, 399-406. https://doi.org/10.3390/i8050399
Zhang Y, Ding J, Wan Y, Xie W, Yuan L. Functional Analysis of the Drosophila Dnop5 Using Targeted RNA Interference. International Journal of Molecular Sciences. 2007; 8(5):399-406. https://doi.org/10.3390/i8050399
Chicago/Turabian StyleZhang, Yan, Jie Ding, Yongqi Wan, Wei Xie, and Liudi Yuan. 2007. "Functional Analysis of the Drosophila Dnop5 Using Targeted RNA Interference" International Journal of Molecular Sciences 8, no. 5: 399-406. https://doi.org/10.3390/i8050399



