Genotoxic Pyrrolizidine Alkaloids — Mechanisms Leading to DNA Adduct Formation and Tumorigenicity
Abstract
:Introduction
Pyrrolizidine alkaloids, widespread plant genotoxicants
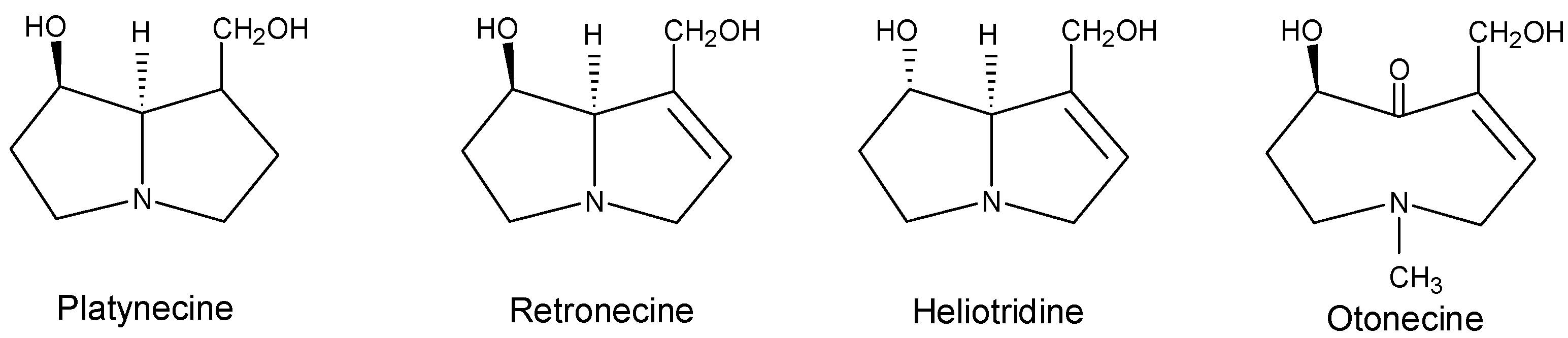
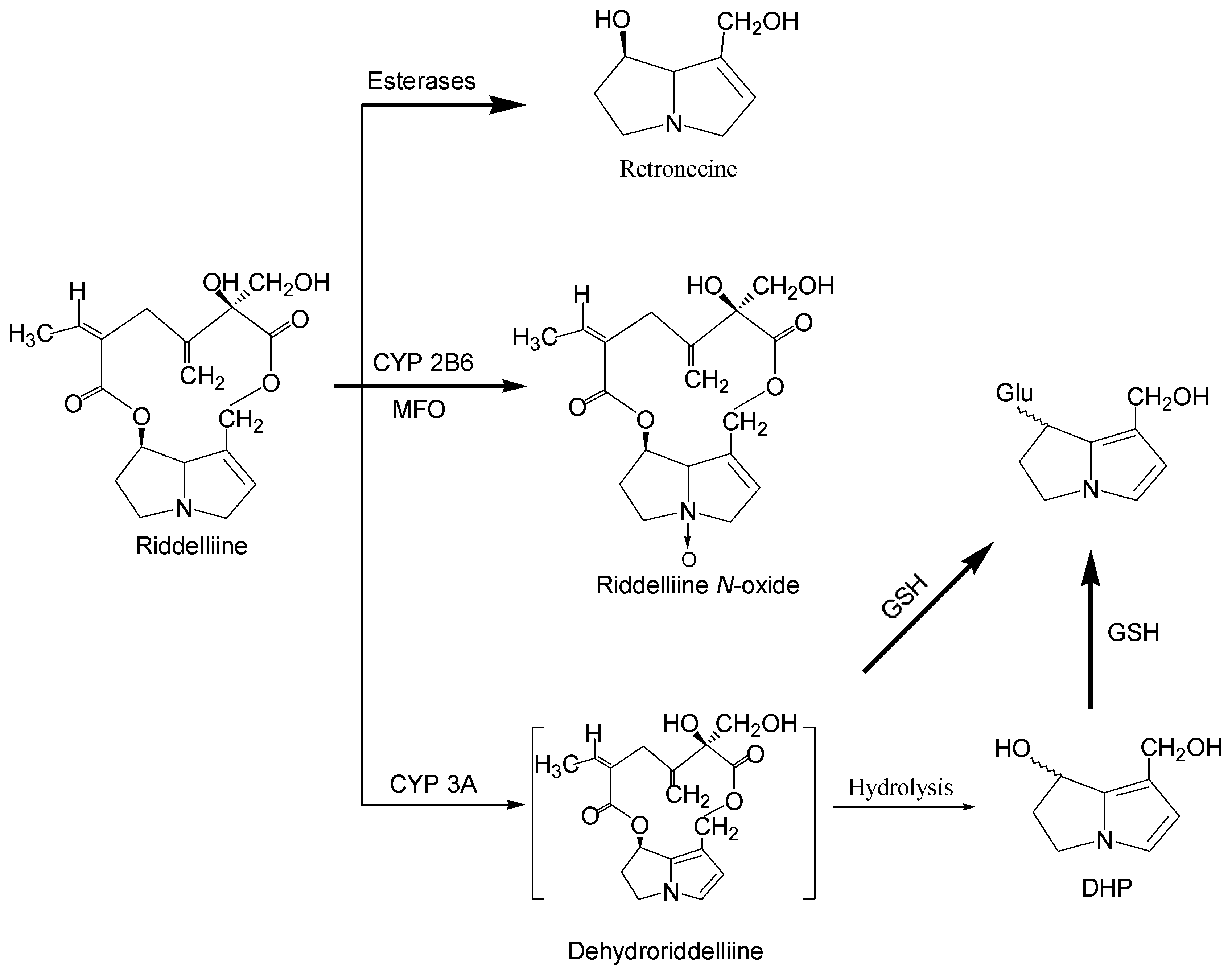
Metabolism and metabolic activation of pyrrolizidine alkaloids
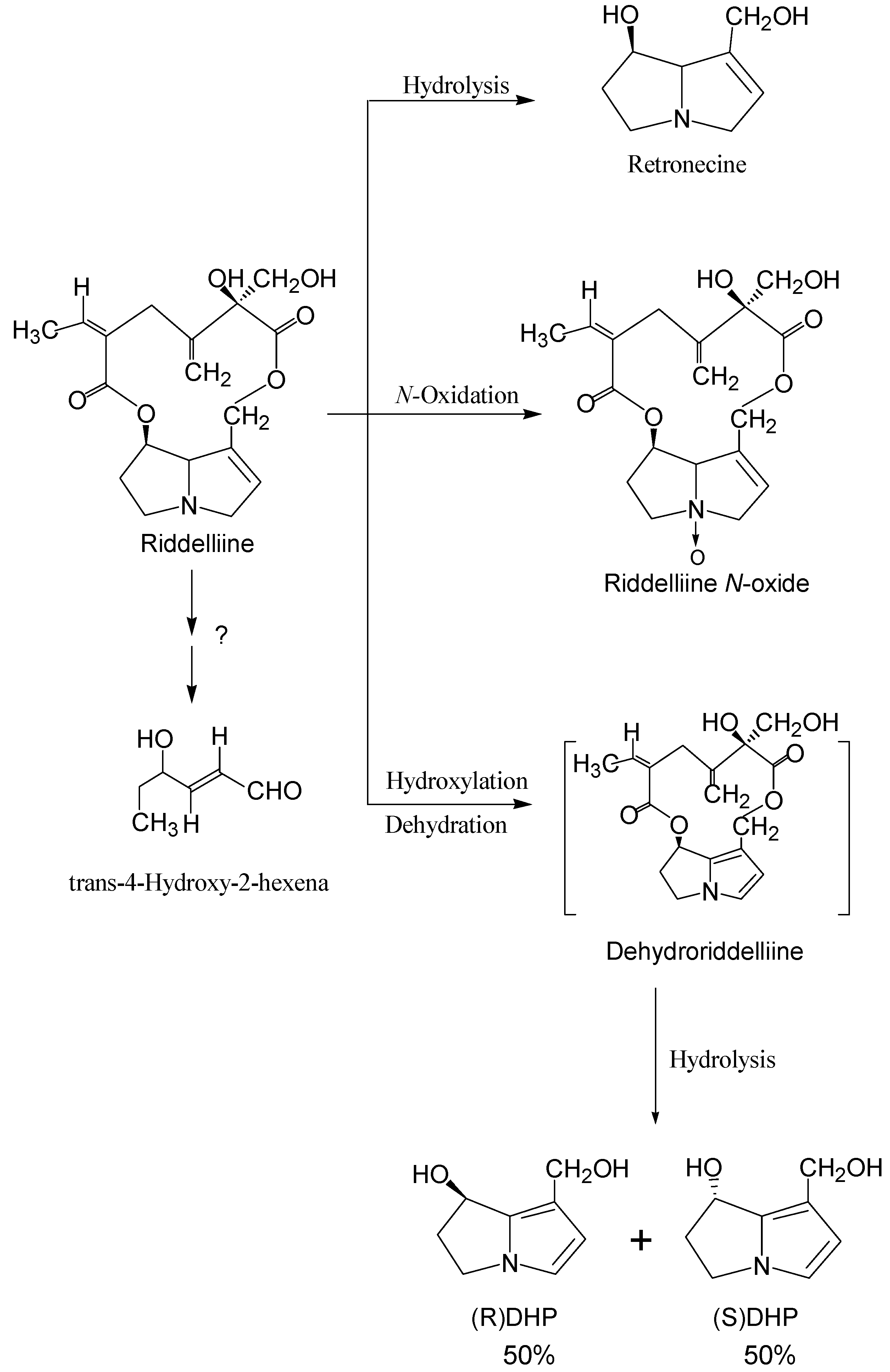
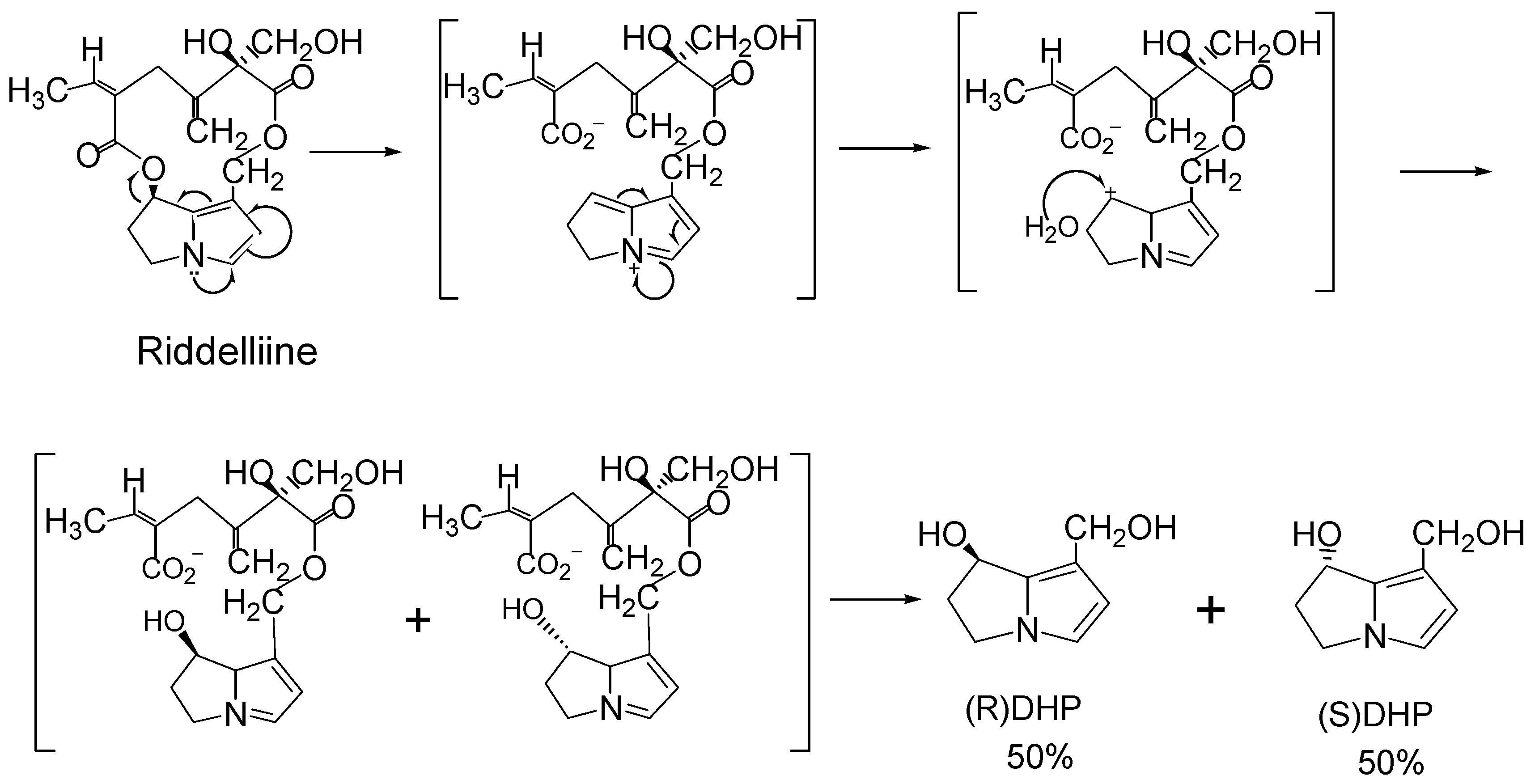
Structure-activity relationship concerning genotoxicity
Tumorigenicity of pyrrolizidine alkaloids
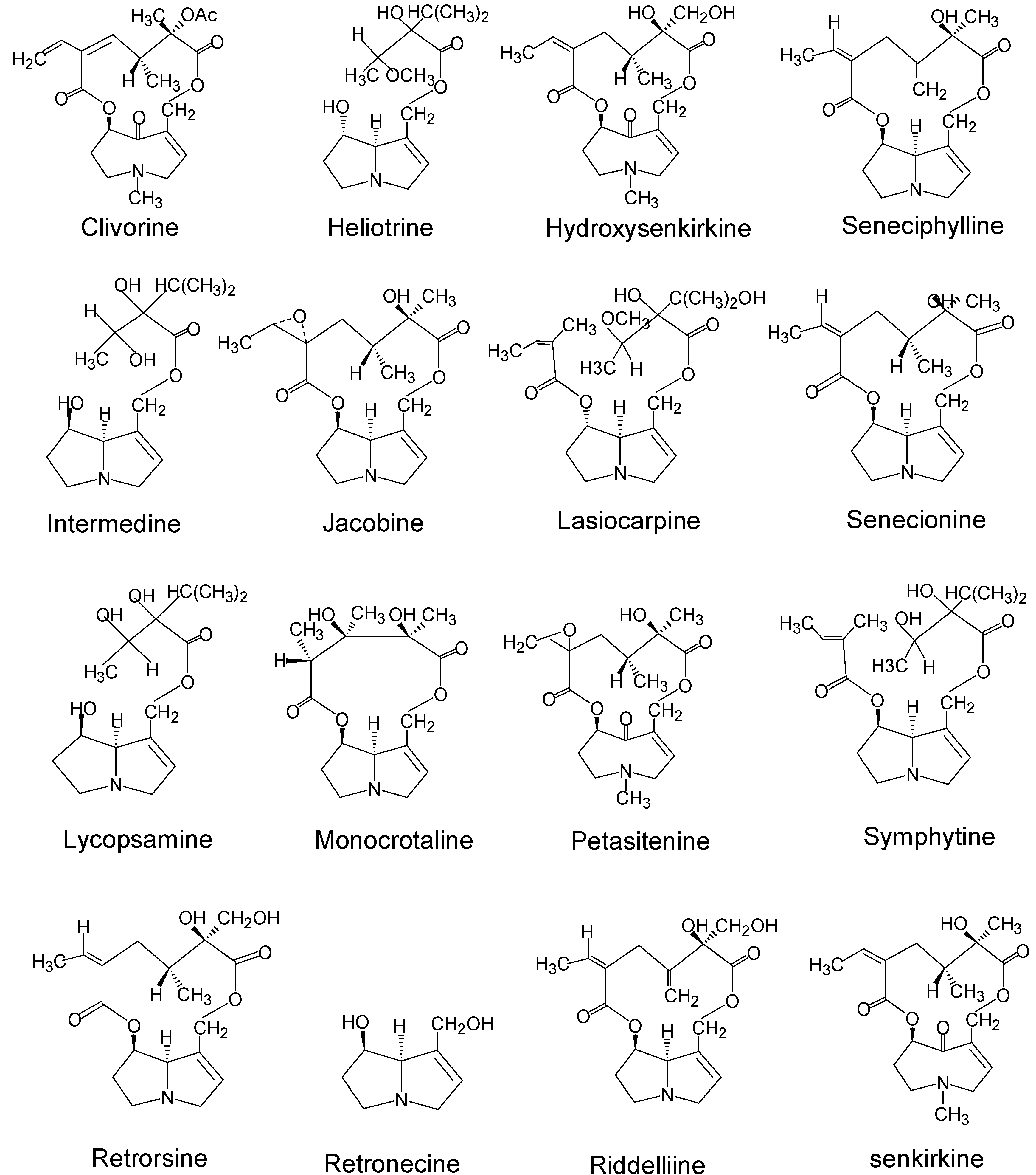
| Pyrrolizidine alkaloids | Plant species (Family)1 | References |
| Clivorine | Ligularia dentata Hara (Compositae) | 28 |
| Heliotrine | Heliotropium (Boraginaceae) | 29 |
| Hydroxy-senkirkine | Senecio (Compositae) | 30,31 |
| Intermedine | Amsinckia (Boraginaceae) | 32,33 |
| Jacobine | Senecio L. (Compositae) | 34,35 |
| Lasiocarpine | Heliotropium (Boraginaceae) | 28,36,37 |
| Lycopasamine | Amsinckia (Boraginaceae) | 32,33 |
| Monocrotaline | Crotalaria (Leguminosae) | 29 |
| Patasitenine | Senecio (Compositae) | 38 |
| Retrorsine | Senecio (Compositae) | 33,34,39 |
| Retronecine | Crotalaria (Leguminosae) | 30 |
| Riddelliine | Senecio (Compositae); Crotalaria (Leguminosae) | 39 |
| Seneciphylline | Senecio (Compositae) | 38,39 |
| Symphytine | Symphytum officinale L (Boraginaceae) | 41,42 |
| Senkirkine | Senecio (Compositae); Petasites (Compositae) | 11,38,41 |
| Senecionine2 | Senecio (Compositae) | 11,34 |
| Isatidine (Retrorsine N-oxide) | Senecio (Compositae), Crotalaria (Leguminosae) | 33,34 |
Mechanisms leading to tumorigenicity
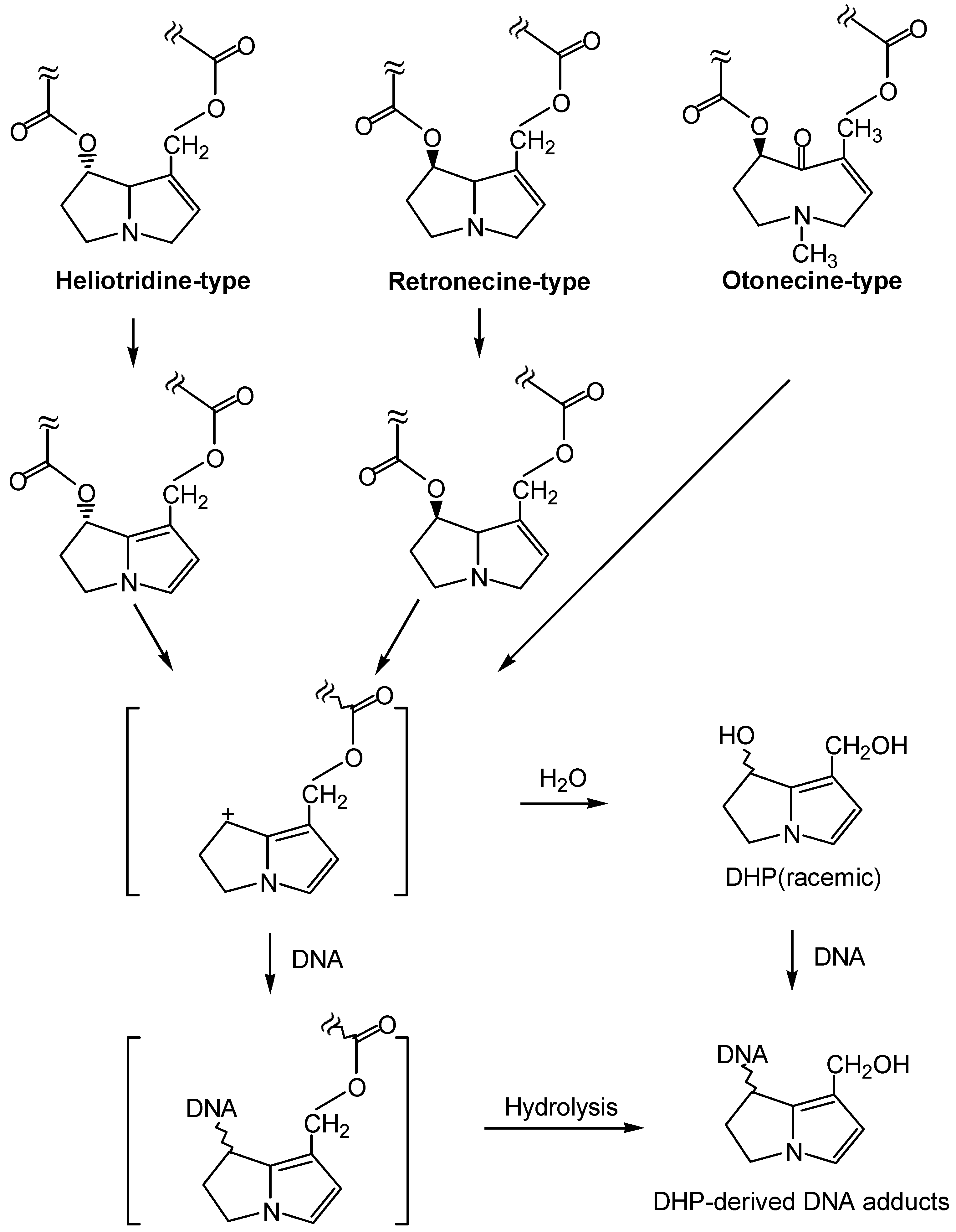
1. Formation of exogenous DNA adducts
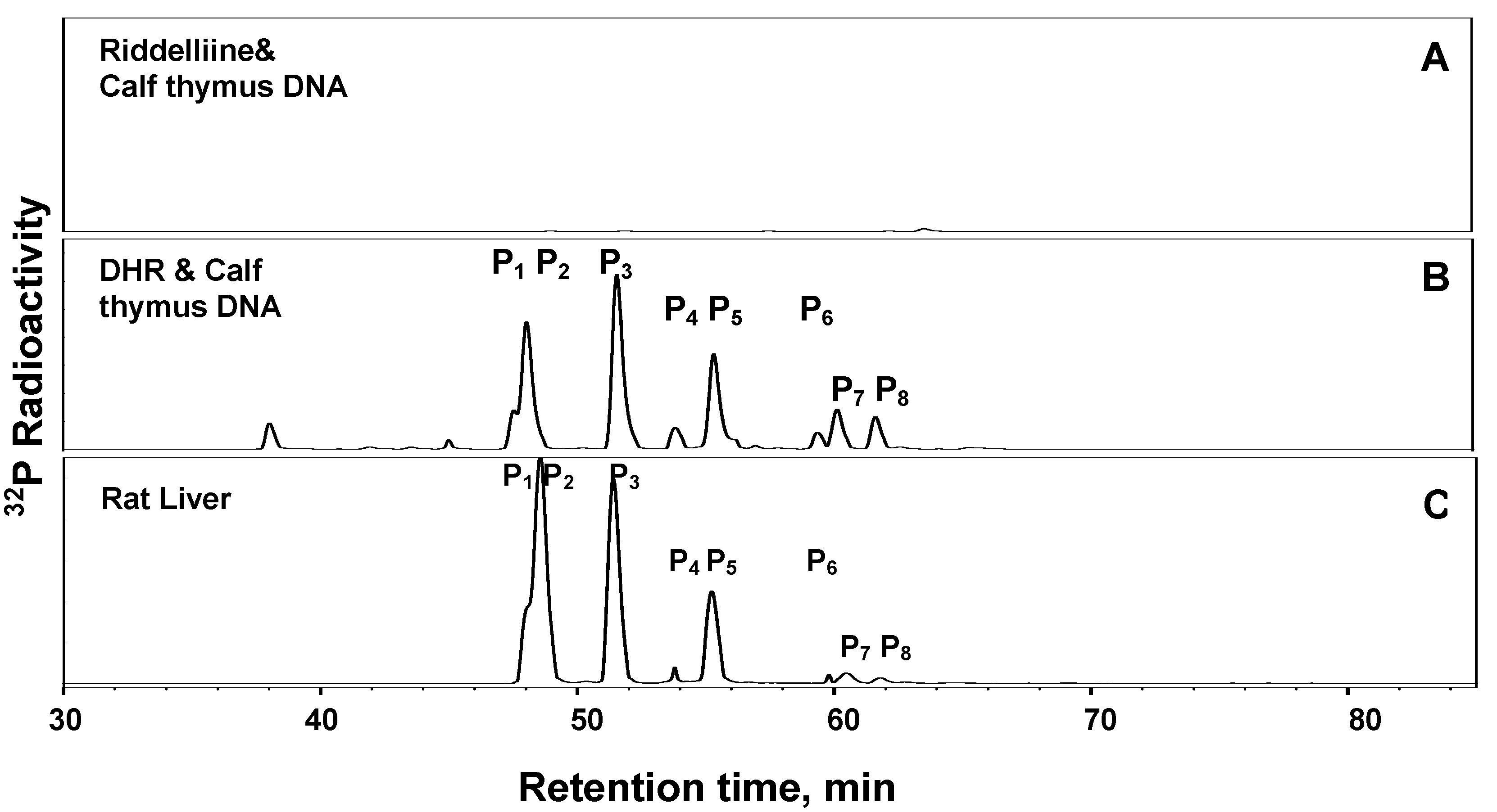
2. Formation of DNA cross-linking and DNA-protein cross-linking
3. Formation of endogenous DNA adducts
Perspectives
References
- International Agency for Research in Cancer (IARC). IARC Monograph on the evaluation of carcinogenic risk of chemicals to man - Some naturally occurring substance; International Agency for Research in Cancer: Lyon, France, 1976. [Google Scholar]
- Mattocks, A. R. Nature 1968, 217, 723–728.
- Mattocks, A. R. Academic Press: London, NY, 1986.
- Roeder, E. Pharmazie 1995, 50, 83–98.
- Stegelmeier, B. L.; Edgar, J. A.; Colegate, S. M.; Gardner, D. R.; Schoch, T. K. J. Nat. Toxins 1999, 8, 95–116.
- Woo, Y.-T.; Lai, D. Y.; Arcos, J. C.; Argus, M. F. Academic Press Inc.: San Diego, 1988.
- Roeder, E. Pharmazie 2000, 55, 711–726.
- Smith, L. W.; Culvenor, C. C. J. Nat. Prod. 1981, 44, 129–152. [CrossRef]
- Betz, J. M.; Eppley, R. M.; Taylor, W. C.; Andrzejewski, D. J. Pharm. Sci. 1994, 83, 649–653. [CrossRef]
- Huxtable, R. J. Biol. Med. 1980, 24, 1–14.
- Hirono, I.; Mori, H.; Culvenor, C. C. Gann 1976, 67, 125–129.
- Winship, K. A. Adverse Drug React. Toxicol. Rev. 1991, 10, 47–59.
- Byron, J. Food Chem. News 1998, 14, 6–7.
- Prakash, A. S.; Pereira, T. N.; Reilly, P. E.; Seawright, A. A. Mutat. Res. 1999, 443, 53–67.
- Yang, Y. C.; Yan, J.; Doerge, D. R.; Chan, P. C.; Fu, P. P. Chem. Res. Toxicol. 2001, 14, 101–109. [CrossRef]
- White, I.N.H.; Mattocks, A.R. Biochem. J. 1972, 128, 291–297.
- Eastman, D.F.; Dimenna, G.P.; Segall, H.J. Drug Metab. Disp. 1982, 10, 236–240.
- Petry, T. W.; Bowden, G. T.; Huxtable, R. J.; Sipes, I. G. Cancer Res. 1984, 44, 1505–1509.
- Fu, P.P.; Chou, M.W.; Xia, Q.; Yang, Y.-C.; Yan, J.; Dorege, D.R.; Chan, P.C. Environ. Carcinogen Ecotoxicol. Rev. 2001, C19 (2), 353.
- Coulombe, R. A., Jr.; Drew, G. L.; Stermitz, F. R. Toxicol. Appl. Pharmacol. 1999, 154, 198–202. [CrossRef]
- Hincks, J. R.; Kim, H. Y.; Segall, H. J.; Molyneux, R. J.; Stermitz, F. R. Toxicol. Appl. Pharmacol. 1991, 111, 90–98. [CrossRef]
- Kim, H. Y.; Stermitz, F. R.; Li, J. K.; Coulombe, R. A. Food Chem. Toxicol. 1999, 37, 619–625.
- Pereira, T. N.; Webb, R. I.; Reilly, P. E.; Seawright, A. A.; Prakash, A. S. Nucleic Acids Res. 1998, 26, 5441–5447.
- Reed, R. L.; Ahern, K. G.; Pearson, G. D.; Buhler, D. R. Carcinogenesis 1988, 9, 1355–1361.
- Hincks, J. R.; Coulombe, R. A. Environ. Mol. Mutag. 1989, 13, 211–217. [CrossRef]
- Kim, H. Y.; Stermitz, F. R.; Coulombe, R. A. Carcinogenesis 1995, 16, 2691–2697.
- Rubiolo, P.; Pieters, L.; Calomme, M.; Bicchi, C.; Vlietinck, A.; et al. Mutat. Res. 1992, 281, 143–147. [CrossRef] [PubMed]
- Kuhara, K.; Takanashi, H.; Hirono, I.; Furuya, T.; Asada, Y. Cancer Lett. 1980, 10, 117–122.
- Schoental, R. Cancer Res. 1975, 35, 2020–2024.
- Schoental, R.; Cavanagh, J.B. J. Natl. Cancer Inst. 1972, 49, 665.
- Crout, D. H. J. Chem. Soc. [Perkin 1] 1972, 13, 1602–1607. [CrossRef]
- Schoental, R.; Fowler, M.E.; Coady, A. Cancer res. 1970, 30, 2127.
- Schoental, R.; Hard, G. C.; Gibbard, S. J. Natl. Cancer Inst. 1971, 47, 1037–1044.
- Schoental, R.; Head, M.A.; Peacock, P.R. Br. J. Cancer 1954, 8, 458. [CrossRef]
- Cook, J. W.; Duffy, E.; Schoental, R. Br. J. Cancer 1950, 4, 405–410. [CrossRef] [Green Version]
- Svoboda, D. J.; Reddy, J. K. Cancer Res. 1972, 32, 908–913.
- Rao, M. S.; Reddy, J. K. Br. J. Cancer 1978, 37, 289–293. [CrossRef]
- Hirono, I.; Ueno, I.; Aiso, S.; Yamaji, T.; Haga, M. Cancer Lett. 1983, 20, 191–198. [CrossRef] [PubMed]
- Harris, P. N.; Chen, K. K. Cancer Res. 1970, 30, 2881–2886.
- Chan, P. C. NIH Publication No. 94-3350. 1993.
- Hirono, I.; Mori, H.; Haga, M. J. Natl. Cancer Inst. 1978, 61, 865–868.
- Hirono, I.; Haga, M.; Fujii, M.; Matsuura, S.; Matsubara, N.; et al. J. Natl. Cancer Inst. 1979, 63, 469–472. [PubMed]
- Brink, N.G. Mutat. Res. 1982, 104, 105.
- Jago, M. V.; Edgar, J. A.; Smith, L. W.; Culvenor, C. C. Mol. Pharmacol. 1970, 6, 402–406.
- Mattocks, A. R.; White, I. N. Chem. Biol. Interact. 1971, 3, 383–396. [CrossRef]
- Eastman, D. F.; Segall, H. J. Toxicol Lett 1981, 8, 217–222.
- Williams, D. E.; Reed, R. L.; Kedzierski, B.; Dannan, G. A.; Guengerich, F. P. Drug Metab. Dispos. 1989, 17, 387–392.
- Miranda, C. L.; Reed, R. L.; Guengerich, F. P.; Buhler, D. R. Carcinogenesis 1991, 12, 515–519.
- Chung, W. G.; Buhler, D. R. Toxicol. Appl. Pharmacol. 1994, 127, 314–319. [CrossRef]
- Reid, M. J.; Lame, M. W.; Morin, D.; Wilson, D. W.; Segall, H. J. J. Biochem. Mol. Toxicol. 1998, 12, 157–166.
- Kasahara, Y.; Kiyatake, K.; Tatsumi, K.; Sugito, K.; Kakusaka, I.; et al. J. Cardiovasc. Pharmacol. 1997, 30, 124–129. [CrossRef]
- Chung, W. G.; Miranda, C. L.; Buhler, D. R. Xenobiotica 1995, 25, 929–939.
- Buhler, D. R.; Kedzierski, B. Adv. Exp. Med. Biol. 1986, 197, 611–620.
- Dueker, S. R.; Lame, M. W.; Morin, D.; Wilson, D. W.; Segall, H. J. Drug Metab. Dispos. 1992, 20, 275–280.
- Lame, M. W.; Jones, A. D.; Morin, D.; Segall, H. J. Drug Metab. Dispos. 1991, 19, 516–524.
- Kedzierski, B.; Buhler, D. R. Toxicol. Lett. 1985, 25, 115–119.
- Kedzierski, B.; Buhler, D. R. Chem. Biol. Interact. 1986, 57, 217–222. [CrossRef]
- Segall, H. J.; Wilson, D. W.; Dallas, J. L.; Haddon, W. F. Science 1985, 229, 472–475.
- Miranda, C. L.; Reed, R. L.; Cheeke, P. R.; Buhler, D. R. Toxicol. Appl. Pharmacol. 1981, 59, 424–430. [CrossRef]
- Lin, G.; Cui, Y. Y.; Hawes, E. M. Drug Metab. Disps. 1998, 26, 181–184.
- Yan, C. C.; Huxtable, R. J. Toxicol. Appl. Pharmacol. 1995, 130, 132–139. [CrossRef]
- Yan, C. C.; Huxtable, R. J. Toxicon 1995, 33, 627–634. [CrossRef] [PubMed]
- White, I. N. H. Chem.-Bio. Interact. 1976, 13, 333–342. [CrossRef]
- Lin, G.; Cui, Y. Y.; Hawes, E. M. Drug Metab. Dispos. 2000, 28, 1475–1483.
- Williams, D. E.; Reed, R. L.; Kedzierski, B.; Ziegler, D. M.; Buhler, D. R. Drug Metab. Dispos. 1989, 17, 380–386.
- Miranda, C. L.; Chung, W.; Reed, R. E.; Zhao, X.; Henderson, M. C. Biochem. Biophys. Res. Commun. 1991, 178, 546–552.
- McLean, E.K. Pharmacol. Rev. 1970, 22, 429.
- Huan, J.-Y.; Miranda, C.L.; Buhler, D.R.; Cheeke, P.R. Toxical. Appl. Pharmacol. 1998, 151, 229. [CrossRef]
- White, I. N. H.; Mattocks, A. R.; Butler, W. H. Chem.-Biol. Interact. 1973, 6, 207–218. [CrossRef] [PubMed]
- Robertson, K. A. Cancer. Res. 1982, 42, 8–14.
- Hincks, J.R.; Kim, H.-Y.; Segall, H.J.; Molyneux, R.J.; Stermitz, F.R.; Coulombe, R.A., jr. Toxicol. Appl. Pharmacol. 1991, 111, 90–98. [CrossRef] [PubMed]
- Kim, H.-Y.; Stermitz, F.R.; Molyneux, R.J.; wilson, D.W.; Taylor, D.; Coulombe, R.A., Jr. Toxicol. Appl. Pharmacol. 1993, 122, 61–69. [CrossRef] [PubMed]
- Yang, Y.; Yan, J.; Churchwell, M.; Beger, R.; Chan, P. Chem. Res. Toxicol. 2001, 14, 91–100. [CrossRef]
- Yan, J.; Yang, Y-C.; Williams, L.; Xia, Q.; Doerge, D. R.; Chou, M. W.; Fu, Peter P. 222nd National Meeting of the American Chemical Society, Chicago, USA, August 26-30, 2001.
- Chou, M. W.; Yan, J.; Nichols, J.; Xia, Q.; Beland, F. A.; Fu, P.P.; Chan, P.C. Proc. Am. Assoc. Cancer Res. 2002, 43, 1718.
- Chou, M. W.; Yan, J.; Nichols, J.; Xia, Q.; Beland, F. A.; Fu, P.P.; Chan, P.C. Cancer Lett. (Submitted).
- Xia, Q.; Chou, M. W.; Kadlubar, F. F.; Chan, Po-Cheun; Fu, P. P. Proc. Am. Assoc. Cancer Res. 2002, 43, 1722.
- Yan, J.; Jasyl Nichols, J.; Yang, Y.-C.; Fu, P.P.; Chou, M.W. Int. J. Mol. Sci. submitted for publication.
- Segall, H.J. Science 1985, 229, 472–475.
© 2002 by MDPI (http://www.mdpi.org).
Share and Cite
Fu, P.P.; Xia, Q.; Lin, G.; Chou, M.W. Genotoxic Pyrrolizidine Alkaloids — Mechanisms Leading to DNA Adduct Formation and Tumorigenicity. Int. J. Mol. Sci. 2002, 3, 948-964. https://doi.org/10.3390/i3090948
Fu PP, Xia Q, Lin G, Chou MW. Genotoxic Pyrrolizidine Alkaloids — Mechanisms Leading to DNA Adduct Formation and Tumorigenicity. International Journal of Molecular Sciences. 2002; 3(9):948-964. https://doi.org/10.3390/i3090948
Chicago/Turabian StyleFu, Peter P., Qingsu Xia, Ge Lin, and Ming W. Chou. 2002. "Genotoxic Pyrrolizidine Alkaloids — Mechanisms Leading to DNA Adduct Formation and Tumorigenicity" International Journal of Molecular Sciences 3, no. 9: 948-964. https://doi.org/10.3390/i3090948




