Latest Trends in Lipase-Catalyzed Synthesis of Ester Carbohydrate Surfactants: From Key Parameters to Opportunities and Future Development
Abstract
:1. Introduction
2. Optimization of Lipase Enzymatic Carbohydrate Ester Synthesis in the 21st Century: Influence of Key Parameters
2.1. Enzyme Selection
2.2. Key Information
2.3. Temperature
2.4. Substrate Molar Ratio
3. Latest Improvements and Recent Trends of the 21st Century
3.1. Recent Developments in Support Immobilization
3.2. Potentiality of Multi-Enzymes
3.3. Chemo-Enzymatic Synthesis
3.4. Interest of Flow Chemistry
3.5. Molecular Bio-Imprinting as a Promising Tool for Simple Improvement
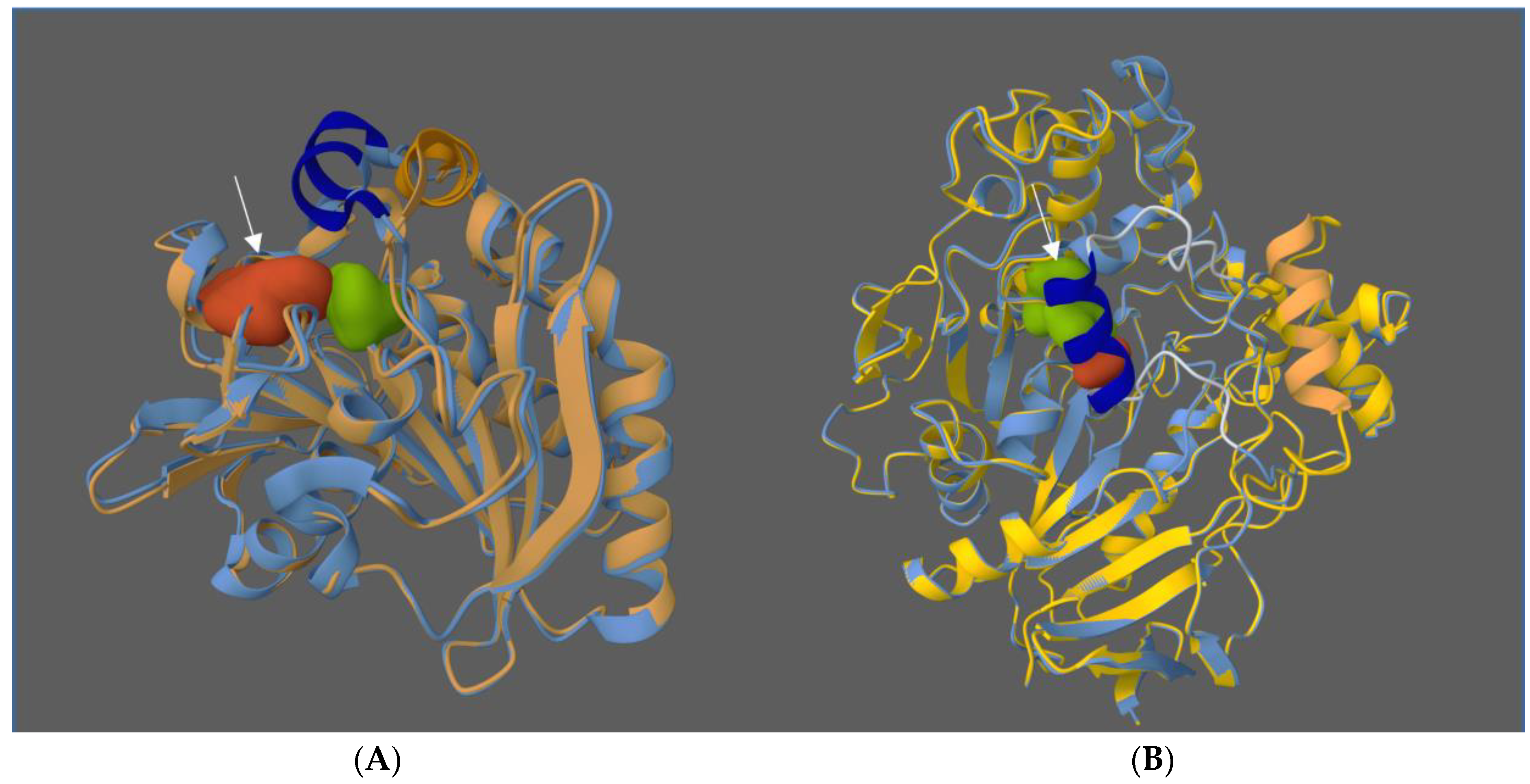
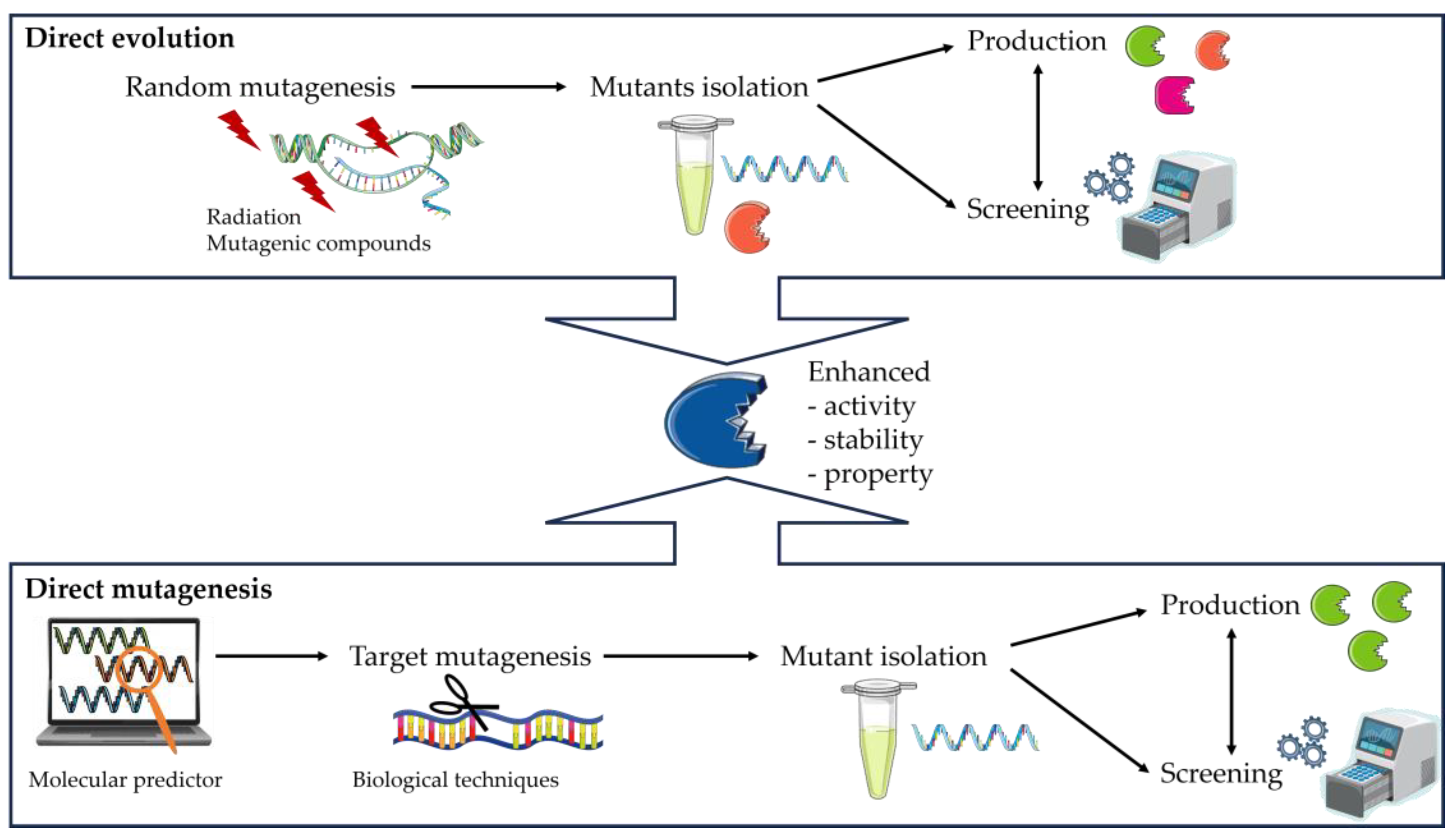
3.6. Enzyme Engineering
- The creation of a library of DNA variants using molecular biology techniques is the first stage.
- The target proteins are then produced using host cells.
- Finally, the enzymes are evaluated using functional screening techniques such as assays to assess the effects of the modifications, such as increased selectivity, reaction specificity, or solvent resistance.
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cameotra, S.S.; Makkar, R.S.; Kaur, J.; Mehta, S.K. Synthesis of Biosurfactants and Their Advantages to Microorganisms and Mankind. Adv. Exp. Med. Biol. 2010, 672, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Kang, J.; Kim, D.-H. Surfactants: Recent Advances and Their Applications. Compos. Commun. 2020, 22, 100537. [Google Scholar] [CrossRef]
- Sachdev, D.P.; Cameotra, S.S. Biosurfactants in Agriculture. Appl. Microbiol. Biotechnol. 2013, 97, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial Biosurfactants Production, Applications and Future Potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.L.B.; Cavalcante, F.T.T.; da Silva Moreira, K.; de Castro Monteiro, R.R.; Rocha, T.G.; da Silva Souza, J.E.; da Fonseca, A.M.; Lopes, A.A.S.; Guimarões, A.P.; de Lima, R.K.C.; et al. Lipases Immobilized onto Nanomaterials as Biocatalysts in Biodiesel Production: Scientific Context, Challenges, and Opportunities. Rev. Virtual Química 2021, 13, 875–891. [Google Scholar] [CrossRef]
- Chrzanowski, Ł.; Ławniczak, Ł.; Czaczyk, K. Why Do Microorganisms Produce Rhamnolipids? World J. Microbiol. Biotechnol. 2012, 28, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, M.; Silva, S.S.E. Recent Food Applications of Microbial Surfactants. Crit. Rev. Food Sci. Nutr. 2018, 58, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Rakshit, A.; Acharjee, A.; Saha, B. Novel Amphiphiles and Their Applications for Different Purposes with Special Emphasis on Polymeric Surfactants. ChemistrySelect 2019, 4, 6978–6995. [Google Scholar] [CrossRef]
- Singh, R.S.; Singh, R.P.; Kennedy, J.F. Immobilization of Yeast Inulinase on Chitosan Beads for the Hydrolysis of Inulin in a Batch System. Int. J. Biol. Macromol. 2017, 95, 87–93. [Google Scholar] [CrossRef]
- Kourmentza, C.; Freitas, F.; Alves, V.; Reis, M.A.M. Microbial Conversion of Waste and Surplus Materials into High-Value Added Products: The Case of Biosurfactants. In Microbial Applications Vol.1: Bioremediation and Bioenergy; Kalia, V.C., Kumar, P., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 29–77. ISBN 978-3-319-52666-9. [Google Scholar]
- Gudiña, E.J.; Rodrigues, L.R. Research and Production of Biosurfactants for the Food Industry. In Bioprocessing for Biomolecules Production; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 125–143. ISBN 978-1-119-43443-6. [Google Scholar]
- Sharma, P.; Melkania, U. Biosurfactant-Enhanced Hydrogen Production from Organic Fraction of Municipal Solid Waste Using Co-Culture of E. Coli and Enterobacter Aerogenes. Bioresour. Technol. 2017, 243, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Naughton, P.J.; Marchant, R.; Naughton, V.; Banat, I.M. Microbial Biosurfactants: Current Trends and Applications in Agricultural and Biomedical Industries. J. Appl. Microbiol. 2019, 127, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Verboni, M.; Lucarini, S.; Duranti, A. 6′-O-Lactose Ester Surfactants as an Innovative Opportunity in the Pharmaceutical Field: From Synthetic Methods to Biological Applications. Pharmaceuticals 2021, 14, 1306. [Google Scholar] [CrossRef] [PubMed]
- Markande, A.; Acharya, S.; Nerurkar, A. Physicochemical Characterization of a Thermostable Glycoprotein Bioemulsifier from Solibacillus Silvestris AM1. Process Biochem. 2013, 48, 1800–1808. [Google Scholar] [CrossRef]
- Nott, K.; Richard, G.; Laurent, P.; Jérôme, C.; Blecker, C.; Wathelet, J.-P.; Paquot, M.; Deleu, M. Enzymatic Synthesis and Surface Properties of Novel Rhamnolipids. Process Biochem. 2013, 48, 133–143. [Google Scholar] [CrossRef]
- Perugino, G. Oligosaccharide Synthesis by Glycosynthases. Trends Biotechnol. 2004, 22, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Gloster, T.M. Exploitation of Carbohydrate Processing Enzymes in Biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Piccicuto, S.; Blecker, C.; Brohée, J.-C.; Mbampara, A.; Lognay, G.; Deroanne, C.; Paquot, M.; Marlier, M. Les esters de sucres: Voies de synthèse et potentialités d’utilisation. Biotechnol. Agron. Soc. Environ. 2001, 5, 209–219. [Google Scholar]
- Deleu, M.; Paquot, M. From Renewable Vegetables Resources to Microorganisms: New Trends in Surfactants. Comptes Rendus Chim. 2004, 7, 641–646. [Google Scholar] [CrossRef]
- Santi, M.; Sancineto, L.; Nascimento, V.; Braun Azeredo, J.; Orozco, E.V.M.; Andrade, L.H.; Gröger, H.; Santi, C. Flow Biocatalysis: A Challenging Alternative for the Synthesis of APIs and Natural Compounds. Int. J. Mol. Sci. 2021, 22, 990. [Google Scholar] [CrossRef]
- Singh, P.; Patil, Y.; Rale, V. Biosurfactant Production: Emerging Trends and Promising Strategies. J. Appl. Microbiol. 2019, 126, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.S.; Alvarado, J.G.; Zambrano, F.; Marquez, R. Surfactants Produced from Carbohydrate Derivatives: A Review of the Biobased Building Blocks Used in Their Synthesis. J. Surfactants Deterg. 2022, 25, 147–183. [Google Scholar] [CrossRef]
- Klein, W.P.; Thomsen, R.P.; Turner, K.B.; Walper, S.A.; Vranish, J.; Kjems, J.; Ancona, M.G.; Medintz, I.L. Enhanced Catalysis from Multienzyme Cascades Assembled on a DNA Origami Triangle. ACS Nano 2019, 13, 13677–13689. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, D.; Isoda, H.; Nakahara, T. Functions and Potential Applications of Glycolipid Biosurfactants—from Energy-Saving Materials to Gene Delivery Carriers. J. Biosci. Bioeng. 2002, 94, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Littlechild, J.A. Improving the “tool Box” for Robust Industrial Enzymes. J. Ind. Microbiol. Biotechnol. 2017, 44, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Fryszkowska, A.; Devine, P.N. Biocatalysis in Drug Discovery and Development. Curr. Opin. Chem. Biol. 2020, 55, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Mateos, A.I.; Contente, M.L.; Roura Padrosa, D.; Paradisi, F. Flow Biocatalysis 101: Design, Development and Applications. React. Chem. Eng. 2021, 6, 599–611. [Google Scholar] [CrossRef]
- Zaks, A.; Klibanov, A.M. Enzyme-Catalyzed Processes in Organic Solvents. J. Biol. Chem. 1985, 82, 3192–3196. [Google Scholar] [CrossRef] [PubMed]
- Torrelo, G.; Hanefeld, U.; Hollmann, F. Biocatalysis. Catal. Lett. 2015, 145, 309–345. [Google Scholar] [CrossRef]
- Çetinkaya, S.; Yenidünya, A.F.; Başoğlu, F.; Saraç, K. D-Glucose-Fatty Acid Ester Synthesis with or without a Biocatalyst in the Same Organic Media. J. Oleo Sci. 2020, 69, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Nikulin, M.; Švedas, V. Prospects of Using Biocatalysis for the Synthesis and Modification of Polymers. Molecules 2021, 26, 2750. [Google Scholar] [CrossRef] [PubMed]
- Barros, E.L.S.; Rebelatto, E.A.; Mayer, D.A.; Wancura, J.H.C.; Oliveira, J.V. Lipase-Catalyzed Production of Sugar Esters in Pressurized Fluid Media: A Review. Chem. Eng. Process.-Process Intensif. 2023, 191, 109480. [Google Scholar] [CrossRef]
- Almeida, F.L.C.; Castro, M.P.J.; Travália, B.M.; Forte, M.B.S. Trends in Lipase Immobilization: Bibliometric Review and Patent Analysis. Process Biochem. 2021, 110, 37–51. [Google Scholar] [CrossRef]
- Chahiniana, H.; Sarda, L. Distinction Between Esterases and Lipases: Comparative Biochemical Properties of Sequence-Related Carboxylesterases. Protein Pept. Lett. 2009, 16, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, J.; Fischer, M.; Schmid, R.D. Anatomy of Lipase Binding Sites: The Scissile Fatty Acid Binding Site. Chem. Phys. Lipids 1998, 93, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Chaput, L.; Marton, Z.; Pineau, P.; Domon, L.; Tran, V.; Graber, M. Enhancing the Enantioselectivity of CALB by Substrate Imprinting: A Combined Experimental and Molecular Dynamics Simulation Model Study. J. Mol. Catal. B Enzym. 2012, 84, 55–61. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Puskas, J. Green Polymer Chemistry: Enzyme Catalysis for Polymer Functionalization. Molecules 2015, 20, 9358–9379. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, Z.S.; Dodson, E.J.; Dodson, G.G.; Turkenburg, J.P. Structure and Molecular Model Refinement of Rhizomucor Miehei Triacyglyceride Lipase: A Case Study of the Use of Simulated Annealing in Partial Model Refinement. Acta Cryst B 1992, 48, 307–319. [Google Scholar] [CrossRef]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the Interface: The Anatomy of a Conformational Change in a Triglyceride Lipase. Biochemistry 1992, 31, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Bouthillier, F.; Smith, P.; Harrison, D.; Rubin, B.; Cygler, M. Insights into Interfacial Activation from an Open Structure of Candida Rugosa Lipase. J. Biol. Chem. 1993, 268, 12843–12847. [Google Scholar] [CrossRef] [PubMed]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Cygler, M. Two Conformational States of Candida Rugosa Lipase. Protein Sci. 1994, 3, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Kourist, R.; Bornscheuer, U.T. Biocatalytic Synthesis of Optically Active Tertiary Alcohols. Appl. Microbiol. Biotechnol. 2011, 91, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Paiva, A.L.; Balcão, V.M.; Malcata, F.X. Kinetics and Mechanisms of Reactions Catalyzed by Immobilized Lipases*. Enzym. Microb. Technol. 2000, 27, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Albayati, S.H.; Masomian, M.; Ishak, S.N.H.; Mohamad Ali, M.S.B.; Thean, A.L.; Mohd Shariff, F.B.; Muhd Noor, N.D.B.; Raja Abd Rahman, R.N.Z. Main Structural Targets for Engineering Lipase Substrate Specificity. Catalysts 2020, 10, 747. [Google Scholar] [CrossRef]
- Hedfors, C.; Hult, K.; Martinelle, M. Lipase Chemoselectivity towards Alcohol and Thiol Acyl Acceptors in a Transacylation Reaction. J. Mol. Catal. B Enzym. 2010, 66, 120–123. [Google Scholar] [CrossRef]
- Chandler, I.C. Determining the Regioselectivity of Immobilized Lipases in Triacylglycerol Acidolysis Reactions. J. Am. Oil Chem. Soc. 2001, 78, 737–742. [Google Scholar] [CrossRef]
- Johnston, J.B.; Singh, A.A.; Clary, A.A.; Chen, C.-K.; Hayes, P.Y.; Chow, S.; De Voss, J.J.; Ortiz de Montellano, P.R. Substrate Analog Studies of the ω-Regiospecificity of Mycobacterium tuberculosis Cholesterol Metabolizing Cytochrome P450 Enzymes CYP124A1, CYP125A1 and CYP142A1. Bioorganic Med. Chem. 2012, 20, 4064–4081. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase Catalysis in Organic Solvents: Advantages and Applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Sang, I.; Park, S.J.; Yoon, H. Enzymatic Synthesis of Sugar Fatty Acid Esters. J. Ind. Eng. Chem. 2007, 13, 1–6. [Google Scholar]
- Gonçalves, M.C.P.; Cansian, A.B.M.; Tardioli, P.W.; Saville, B.A. Production of Sugars from Mixed Hardwoods for Use in the Synthesis of Sugar Fatty Acid Esters Catalyzed by Immobilized-Stabilized Derivatives of Candida Antarctica Lipase B. Biofuels Bioprod. Biorefin. 2023, 17, 1236–1250. [Google Scholar] [CrossRef]
- Afach, G.; Kawanami, Y.; Cheetangdee, N.; Fukada, K.; Izumori, K. Lipase-Catalyzed Synthesis of d-Psicose Fatty Acid Diesters and Their Emulsification Activities. J. Am. Oil Chem. Soc. 2008, 85, 755–760. [Google Scholar] [CrossRef]
- Buzatu, A.R.; Soler, M.A.; Fortuna, S.; Ozkilinc, O.; Dreavă, D.M.; Bîtcan, I.; Badea, V.; Giannozzi, P.; Fogolari, F.; Gardossi, L.; et al. Reactive Natural Deep Eutectic Solvents Increase Selectivity and Efficiency of Lipase Catalyzed Esterification of Carbohydrate Polyols. Catal. Today 2024, 426, 114373. [Google Scholar] [CrossRef]
- Khaled Khodja, N.; Montet, D.; Pina, M.; Graille, J. Fructose Oleate Synthesis in a Fixed Catalyst Bed Reactor. Biotechnol. Lett. 1991, 13, 167–172. [Google Scholar] [CrossRef]
- Li, L.; Ji, F.; Wang, J.; Jiang, B.; Li, Y.; Bao, Y. Efficient Mono-Acylation of Fructose by Lipase-Catalyzed Esterification in Ionic Liquid Co-Solvents. Carbohydr. Res. 2015, 416, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Gumel, A.M.; Annuar, M.S.M.; Heidelberg, T.; Chisti, Y. Lipase Mediated Synthesis of Sugar Fatty Acid Esters. Process Biochem. 2011, 46, 2079–2090. [Google Scholar] [CrossRef]
- Cabezas, J.T.; Waglay, A.; Karboune, S. Lipase-Catalyzed Synthesis of Fructosyl Myristic Acid Esters as Biosurfactants in a Low Solvent Media: Optimization of the Bioconversion. Food Biosci. 2022, 50, 102026. [Google Scholar] [CrossRef]
- Abdulmalek, E.; Saupi, H.S.M.; Tejo, B.A.; Basri, M.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Rahman, M.B.A. Improved Enzymatic Galactose Oleate Ester Synthesis in Ionic Liquids. J. Mol. Catal. B Enzym. 2012, 76, 37–43. [Google Scholar] [CrossRef]
- Lin, X.-S.; Zhao, K.-H.; Zhou, Q.-L.; Xie, K.-Q.; Halling, P.J.; Yang, Z. Aspergillus Oryzae Lipase-Catalyzed Synthesis of Glucose Laurate with Excellent Productivity. Bioresour. Bioprocess. 2016, 3, 2. [Google Scholar] [CrossRef]
- Cauglia, F.; Canepa, P. The Enzymatic Synthesis of Glucosylmyristate as a Reaction Model for General Considerations on ‘Sugar Esters’ Production. Bioresour. Technol. 2008, 99, 4065–4072. [Google Scholar] [CrossRef]
- Woudenberg-van Oosterom, M.; van Rantwijk, F.; Sheldon, R.A. Regioselective Acylation of Disaccharides in Tert-Butyl Alcohol Catalyzed by Candida Antarctica Lipase. Biotechnol. Bioeng. 1996, 49, 328–333. [Google Scholar] [CrossRef]
- Enayati, M.; Gong, Y.; Goddard, J.M.; Abbaspourrad, A. Synthesis and Characterization of Lactose Fatty Acid Ester Biosurfactants Using Free and Immobilized Lipases in Organic Solvents. Food Chem. 2018, 266, 508–513. [Google Scholar] [CrossRef] [PubMed]
- García-Oliva, C.; Perona, A.; Rumbero, Á.; Hoyos, P.; Hernáiz, M.J. Enzymatic Synthesis and Molecular Modelling Studies of Rhamnose Esters Using Lipase from Pseudomonas Stutzeri. Int. J. Mol. Sci. 2022, 23, 2239. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Alfonso, J.L.; Casas-Godoy, L.; Arrizon, J.; Arrieta-Baez, D.; Ballesteros, A.O.; Sandoval, G.; Plou, F.J. Lipase-Catalyzed Synthesis of Fatty Acid Esters of Trisaccharides. In Lipases and Phospholipases: Methods and Protocols; Sandoval, G., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 287–296. ISBN 978-1-4939-8672-9. [Google Scholar]
- Delavault, A.; Ochsenreither, K.; Syldatk, C. Intensification of Biocatalytic Processes by Using Alternative Reaction Media. Phys. Sci. Rev. 2023. [Google Scholar] [CrossRef]
- Zago, E.; Joly, N.; Chaveriat, L.; Lequart, V.; Martin, P. Enzymatic Synthesis of Amphiphilic Carbohydrate Esters: Influence of Physicochemical and Biochemical Parameters. Biotechnol. Rep. 2021, 30, e00631. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Yamada, R.; Ogino, H. Chemical Treatments for Modification and Immobilization to Improve the Solvent-Stability of Lipase. World J. Microbiol. Biotechnol. 2019, 35, 193. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.R.; Silva, A.S.; Fernandez-Lafuente, R.; Ferreira-Leitão, V.S. Solvent-Free Esterifications Mediated by Immobilized Lipases: A Review from Thermodynamic and Kinetic Perspectives. Catal. Sci. Technol. 2021, 11, 5696–5711. [Google Scholar] [CrossRef]
- Wehtje, E.; Costes, D.; Adlercreutz, P. Enantioselectivity of Lipases: Effects of Water Activity. J. Mol. Catal. B Enzym. 1997, 3, 221–230. [Google Scholar] [CrossRef]
- Ducret, A.; Trani, M.; Lortie, R. Lipase-Catalyzed Enantioselective Esterification of Ibuprofen in Organic Solvents under Controlled Water Activity. Enzym. Microb. Technol. 1998, 22, 212–216. [Google Scholar] [CrossRef]
- Gonçalves, M.C.P.; Romanelli, J.P.; Guimarães, J.R.; Vieira, A.C.; de Azevedo, B.P.; Tardioli, P.W. Reviewing Research on the Synthesis of CALB-Catalyzed Sugar Esters Incorporating Systematic Mapping Principles. Crit. Rev. Biotechnol. 2021, 41, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Hollenbach, R.; Ochsenreither, K.; Syldatk, C. Enzymatic Synthesis of Glucose Monodecanoate in a Hydrophobic Deep Eutectic Solvent. Int. J. Mol. Sci. 2020, 21, 4342. [Google Scholar] [CrossRef] [PubMed]
- Bouzaouit, N.; Bidjou-Haiour, C. Response Surface Methodological Study of Glucose Laurate Synthesis Catalyzed by Immobilized Lipase from Candida Cylindracea. Biol. Forum. 2016, 8, 420. [Google Scholar]
- Hollenbach, R.; Ochsenreither, K.; Syldatk, C. Parameters Influencing Lipase-Catalyzed Glycolipid Synthesis by (Trans-)Esterification Reaction. Adv. Biochem. Eng./Biotechnol. 2022, 181, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Dordick, J.S.; Garde, S. Hydration of Enzyme in Nonaqueous Media Is Consistent with Solvent Dependence of Its Activity. Biophys. J. 2004, 87, 812–821. [Google Scholar] [CrossRef]
- Idris, A.; Bukhari, A. Immobilized Candida antarctica Lipase B: Hydration, Stripping off and Application in Ring Opening Polyester Synthesis. Biotechnol. Adv. 2012, 30, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Trodler, P.; Pleiss, J. Modeling Structure and Flexibility of Candida Antarctica Lipase B in Organic Solvents. BMC Struct. Biol. 2008, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Castillo, E.; Torres-Gavilán, A.; Sandoval, G.; Marty, A. Thermodynamical Methods for the Optimization of Lipase-Catalyzed Reactions. Methods Mol. Biol. 2012, 861, 383–400. [Google Scholar] [CrossRef] [PubMed]
- Degn, P.; Zimmermann, W. Optimization of Carbohydrate Fatty Acid Ester Synthesis in Organic Media by a Lipase from Candida Antarctica. Biotechnol. Bioeng. 2001, 74, 483–491. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, J.; Feng, B.; Zhang, X.; Xia, W. A Simple Approach for the Selective Enzymatic Synthesis of Dilauroyl Maltose in Organic Media. J. Mol. Catal. B Enzym. 2010, 62, 265–269. [Google Scholar] [CrossRef]
- Cao, L.; Fischer, A.; Bornscheuer, U.T.; Schmid, R.D. Lipase-Catalyzed Solid Phase Synthesis of Sugar Fatty Acid Esters. Biocatal. Biotransform. 1996, 14, 269–283. [Google Scholar] [CrossRef]
- Hansch, C.; Leo, A.; Hoekman, D.H. Exploring QSAR: Hydrophobic, Electronic, and Steric Constants; American Chemical Society: Washington, DC, USA, 1995; Volume 2, ISBN 978-0-8412-2991-4. [Google Scholar]
- O’Neil, M.J. (Ed.) The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 15th ed.; Royal Society of Chemistry: Cambridge, UK, 2013; ISBN 978-1-84973-670-1. [Google Scholar]
- Haynes, W.M.; Lide, D.R.; Bruno, T.J. (Eds.) CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data, 94th ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2013; ISBN 978-1-4665-7114-3. [Google Scholar]
- O’Neil, M.J. The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 13th ed.; Merck: Whitehouse Station, NJ, USA, 2001; ISBN 978-0-911910-13-1. [Google Scholar]
- Shin, D.W.; Mai, N.L.; Bae, S.-W.; Koo, Y.-M. Enhanced Lipase-Catalyzed Synthesis of Sugar Fatty Acid Esters Using Supersaturated Sugar Solution in Ionic Liquids. Enzym. Microb. Technol. 2019, 126, 18–23. [Google Scholar] [CrossRef]
- Degn, P.; Pedersen, L.H.; Duus, J.Ø.; Zimmermann, W. Lipase Catalyzed Synthesis of Glucose Fatty Acid Esters in Tert-Butanol. Biotechnol. Lett. 1999, 21, 275–280. [Google Scholar] [CrossRef]
- Martin-Arjol, I.; Isbell, T.; Manresa, A. Mono-Estolide Synthesis from Trans-8-Hydroxy-Fatty Acids by Lipases in Solvent-Free Media and Their Physical Properties. J. Am. Oil Chem. Soc. 2015, 92, 1125–1141. [Google Scholar] [CrossRef]
- Tai, H.P.; Brunner, G. Sugar Fatty Acid Ester Synthesis in High-Pressure Acetone–CO2 System. J. Supercrit. Fluids 2009, 48, 36–40. [Google Scholar] [CrossRef]
- Hidayat, C.; Fitria, K.; Supriyanto; Hastuti, P. Enzymatic Synthesis of Bio-Surfactant Fructose Oleic Ester Using Immobilized Lipase on Modified Hydrophobic Matrix in Fluidized Bed Reactor. Agric. Agric. Sci. Procedia 2016, 9, 353–362. [Google Scholar] [CrossRef]
- Ye, R.; Pyo, S.-H.; Hayes, D.G. Lipase-Catalyzed Synthesis of Saccharide–Fatty Acid Esters Using Suspensions of Saccharide Crystals in Solvent-Free Media. J. Am. Oil Chem. Soc. 2010, 87, 281–293. [Google Scholar] [CrossRef]
- Šabeder, S.; Habulin, M.; Knez, Ž. Lipase-Catalyzed Synthesis of Fatty Acid Fructose Esters. J. Food Eng. 2006, 77, 880–886. [Google Scholar] [CrossRef]
- Vuillemin, M.E.; Husson, E.; Laclef, S.; Jamali, A.; Lambertyn, V.; Pilard, S.; Cailleu, D.; Sarazin, C. Improving the Environmental Compatibility of Enzymatic Synthesis of Sugar-Based Surfactants Using Green Reaction Media. Process Biochem. 2022, 117, 30–41. [Google Scholar] [CrossRef]
- Spalletta, A.; Joly, N.; Martin, P. Optimization of Enzymatic Synthesis of D-Glucose-Based Surfactants Using Supported Aspergillus Niger Lipase as Biocatalyst. Chemistry 2023, 5, 1855–1869. [Google Scholar] [CrossRef]
- Ren, K.; Lamsal, B.P. Synthesis of Some Glucose-Fatty Acid Esters by Lipase from Candida antarctica and Their Emulsion Functions. Food Chem. 2017, 214, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Arcens, D.; Grau, E.; Grelier, S.; Cramail, H.; Peruch, F. 6-O-Glucose Palmitate Synthesis with Lipase: Investigation of Some Key Parameters. Mol. Catal. 2018, 460, 63–68. [Google Scholar] [CrossRef]
- Zaidan, U.H.; Abdul Rahman, M.B.; Othman, S.S.; Basri, M.; Abdulmalek, E.; Abdul Rahman, R.N.Z.R.; Salleh, A.B. Biocatalytic Production of Lactose Ester Catalysed by Mica-Based Immobilised Lipase. Food Chem. 2012, 131, 199–205. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Hayat, K.; Zhang, X.; Shabbar, A.; Feng, B.; Jia, C. Efficient Lipase-Selective Synthesis of Dilauryl Mannoses by Simultaneous Reaction–Extraction System. Biotechnol. Lett. 2009, 31, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Galonde, N.; Nott, K.; Richard, G.; Debuigne, A.; Nicks, F.; Jerome, C.; Fauconnier, M.-L. Study of the Influence of Pure Ionic Liquids on the Lipase-Catalyzed (Trans)Esterification of Mannose Based on Their Anion and Cation Nature. Curr. Org. Chem. 2012, 17, 763–770. [Google Scholar] [CrossRef]
- Chamouleau, F.; Coulon, D.; Girardin, M.; Ghoul, M. Influence of Water Activity and Water Content on Sugar Esters Lipase-Catalyzed Synthesis in Organic Media. J. Mol. Catal. B Enzym. 2001, 11, 949–954. [Google Scholar] [CrossRef]
- Abdulmalek, E.; Hamidon, N.F.; Abdul Rahman, M.B. Optimization and Characterization of Lipase Catalysed Synthesis of Xylose Caproate Ester in Organic Solvents. J. Mol. Catal. B Enzym. 2016, 132, 1–4. [Google Scholar] [CrossRef]
- Villeneuve, P. Lipases in Lipophilization Reactions. Biotechnol. Adv. 2007, 25, 515–536. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Zhang, X.; Liang, F.; Xian, M.; Feng, D.; Ye, Z. Synthesis, Surface Properties of Glucosyl Esters from Renewable Materials for Use as Biosurfactants. Colloids Surf. A Physicochem. Eng. Asp. 2019, 577, 257–264. [Google Scholar] [CrossRef]
- Tsavas, P.; Polydorou, S.; Faflia, I.; Voutsas, E.; Tassios, D.; Flores, M.; Naraghi, K.; Halling, P.; Chamouleau, F.; Ghoul, M.; et al. Solubility of Glucose in Mixtures Containing 2Methyl2-Butanol, Dimethyl Sulfoxide, Acids, Esters, and Water. J. Chem. Eng. Data 2002, 47, 807–810. [Google Scholar] [CrossRef]
- García, C.; Hoyos, P.; Hernáiz, M.J. Enzymatic Synthesis of Carbohydrates and Glycoconjugates Using Lipases and Glycosidases in Green Solvents. Biocatal. Biotransform. 2018, 36, 131–140. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chong, G.H. Chapter 2—Supercritical Fluids (SCFs) as Solvents in the Pharmaceutical Industry. In Green Sustainable Process for Chemical and Environmental Engineering and Science; Inamuddin Boddula, R., Ahamed, M.I., Asiri, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 13–39. ISBN 978-0-12-821885-3. [Google Scholar]
- Sorour, N.; Karboune, S.; Saint-Louis, R.; Kermasha, S. Lipase-Catalyzed Synthesis of Structured Phenolic Lipids in Solvent-Free System Using Flaxseed Oil and Selected Phenolic Acids as Substrates. J. Biotechnol. 2012, 158, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Méline, T.; Muzard, M.; Deleu, M.; Rakotoarivonina, H.; Plantier-Royon, R.; Rémond, C. D-Xylose and L-Arabinose Laurate Esters: Enzymatic Synthesis, Characterization and Physico-Chemical Properties. Enzym. Microb. Technol. 2018, 112, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Aljawish, A.; Heuson, E.; Bigan, M.; Froidevaux, R. Lipase Catalyzed Esterification of Formic Acid in Solvent and Solvent-Free Systems. Biocatal. Agric. Biotechnol. 2019, 20, 101221. [Google Scholar] [CrossRef]
- Otero, C.; Arcos, J.; Berrendero, M.A.; Torres, C. Emulsifiers from Solid and Liquid Polyols: Different Strategies for Obtaining Optimum Conversions and Selectivities. J. Mol. Catal. B Enzym. 2001, 11, 883–892. [Google Scholar] [CrossRef]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.-N.; Dou, Y. Deep Eutectic Solvents for Biocatalytic Transformations: Focused Lipase-Catalyzed Organic Reactions. Appl. Microbiol. Biotechnol. 2020, 104, 1481–1496. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, A.R.; de Maria, P.D. Recent Advances on the Use of 2-Methyltetrahydrofuran (2-MeTHF) in Biotransformations. Curr. Green. Chem. 2018, 5, 86–103. [Google Scholar] [CrossRef]
- Lei, P.; Wang, Y.; Mu, Y.; Wang, Y.; Ma, Z.; Feng, J.; Liu, X.; Szostak, M. Green-Solvent Selection for Acyl Buchwald–Hartwig Cross-Coupling of Amides (Transamidation). ACS Sustain. Chem. Eng. 2021, 9, 14937–14945. [Google Scholar] [CrossRef]
- Donzella, S.; Contente, M.L. The Joint Effort of Enzyme Technology and Flow Chemistry to Bring Biocatalytic Processes to the next Level of Sustainability, Efficiency and Productivity. J. Flow. Chem. 2023. [Google Scholar] [CrossRef]
- Bezbradica, D.; Mijin, D.; Šiler-Marinković, S.; Knežević, Z. The Effect of Substrate Polarity on the Lipase-Catalyzed Synthesis of Aroma Esters in Solvent-Free Systems. J. Mol. Catal. B Enzym. 2007, 45, 97–101. [Google Scholar] [CrossRef]
- Reyes-Duarte, D.; Polaina, J.; López-Cortés, N.; Alcalde, M.; Plou, F.J.; Elborough, K.; Ballesteros, A.; Timmis, K.N.; Golyshin, P.N.; Ferrer, M. Conversion of a Carboxylesterase into a Triacylglycerol Lipase by a Random Mutation. Angew. Chem. (Int. Ed. Engl.) 2005, 44, 7553–7557. [Google Scholar] [CrossRef] [PubMed]
- Jintian, L.; Wentian, Z.; Pingjia, Y.; Yuanan, W. Lipase-Catalyzed Regioselective Synthesis of Palmitolyglucose Ester in Ionic Liquids. Adv. Biol. Chem. 2012, 2012, 226. [Google Scholar] [CrossRef]
- Zhao, K.-H.; Cai, Y.-Z.; Lin, X.-S.; Xiong, J.; Halling, P.J.; Yang, Z. Enzymatic Synthesis of Glucose-Based Fatty Acid Esters in Bisolvent Systems Containing Ionic Liquids or Deep Eutectic Solvents. Molecules 2016, 21, 1294. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, H.; Shaheen, U.; Kadam, T. Characterization of a Hyperthermostable Alkaline Lipase from Bacillus sonorensis 4R. Enzym. Res. 2016, 2016, e4170684. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.H.; Maiangwa, J.; Ali, M.S.M.; Normi, Y.M.; Sabri, S.; Leow, T.C. Thermostable Lipases and Their Dynamics of Improved Enzymatic Properties. Appl. Microbiol. Biotechnol. 2021, 105, 7069–7094. [Google Scholar] [CrossRef]
- Cao, L.; Bornscheuer, U.T.; Schmid, R.D. Lipase-Catalyzed Solid-Phase Synthesis of Sugar Esters. Influence of Immobilization on Productivity and Stability of the Enzyme. J. Mol. Catal. B Enzym. 1999, 6, 279–285. [Google Scholar] [CrossRef]
- Sebatini, A.M.; Jain, M.; Radha, P.; Kiruthika, S.; Tamilarasan, K. Immobilized Lipase Catalyzing Glucose Stearate Synthesis and Their Surfactant Properties Analysis. 3 Biotech 2016, 6, 184. [Google Scholar] [CrossRef]
- Casas-Godoy, L.; Arrizon, J.; Arrieta-Baez, D.; Plou, F.J.; Sandoval, G. Synthesis and Emulsifying Properties of Carbohydrate Fatty Acid Esters Produced from Agave Tequilana Fructans by Enzymatic Acylation. Food Chem. 2016, 204, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Federsel, H.-J.; Moody, T.S.; Taylor, S.J.C. Recent Trends in Enzyme Immobilization—Concepts for Expanding the Biocatalysis Toolbox. Molecules 2021, 26, 2822. [Google Scholar] [CrossRef] [PubMed]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of Lipases on Hydrophobic Supports Involves the Open Form of the Enzyme. Enzym. Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Bonet-Ragel, K.; López-Pou, L.; Tutusaus, G.; Benaiges, M.D.; Valero, F. Rice Husk Ash as a Potential Carrier for the Immobilization of Lipases Applied in the Enzymatic Production of Biodiesel. Biocatal. Biotransform. 2018, 36, 151–158. [Google Scholar] [CrossRef]
- Brígida, A.I.S.; Pinheiro, Á.D.T.; Ferreira, A.L.O.; Gonçalves, L.R.B. Immobilization of Candida Antarctica Lipase B by Adsorption to Green Coconut Fiber. Appl. Biochem. Biotechnol. 2008, 146, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Chu, T.; Chu, J.; Gao, B.; He, B. A Versatile Approach for Enzyme Immobilization Using Chemically Modified 3D-Printed Scaffolds. ACS Sustain. Chem. Eng. 2019, 7, 18048–18054. [Google Scholar] [CrossRef]
- Schultz, N.; Syldatk, C.; Franzreb, M.; Hobley, T.J. Integrated Processing and Multiple Re-Use of Immobilised Lipase by Magnetic Separation Technology. J. Biotechnol. 2007, 132, 202–208. [Google Scholar] [CrossRef]
- Gennari, A.; Führ, A.J.; Volpato, G.; Volken de Souza, C.F. Magnetic Cellulose: Versatile Support for Enzyme Immobilization—A Review. Carbohydr. Polym. 2020, 246, 116646. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, X.; Zheng, R.; Zheng, Y. Immobilization of Multi-Enzymes on Support Materials for Efficient Biocatalysis. Front. Bioeng. Biotechnol. 2020, 8, 660. [Google Scholar] [CrossRef]
- Kong, H. Designing Alginate Hydrogels to Maintain Viability of Immobilized Cells. Biomaterials 2003, 24, 4023–4029. [Google Scholar] [CrossRef] [PubMed]
- Raulo, R.; Heuson, E.; Froidevaux, R.; Phalip, V. Combining Analytical Approaches for Better Lignocellulosic Biomass Degradation: A Way of Improving Fungal Enzymatic Cocktails? Biotechnol. Lett. 2021, 43, 2283–2298. [Google Scholar] [CrossRef] [PubMed]
- Adsul, M.; Sandhu, S.K.; Singhania, R.R.; Gupta, R.; Puri, S.K.; Mathur, A. Designing a Cellulolytic Enzyme Cocktail for the Efficient and Economical Conversion of Lignocellulosic Biomass to Biofuels. Enzym. Microb. Technol. 2020, 133, 109442. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Semwal, S.; Kumar, R.; Mathur, A.; Gupta, R.P.; Tuli, D.K.; Satlewal, A. Synergistic Enzyme Cocktail to Enhance Hydrolysis of Steam Exploded Wheat Straw at Pilot Scale. Front. Energy Res. 2018, 6, 122. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, H.; Yan, Y. Optimization of Lipase-Catalyzed Transesterification of Lard for Biodiesel Production Using Response Surface Methodology. Appl. Biochem. Biotechnol. 2010, 160, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Arcens, D.; Fer, G.L.; Grau, E.; Grelier, S.; Cramail, H.; Peruch, F. Chemo-Enzymatic Synthesis of Glycolipids, Their Polymerization and Self-Assembly. Polym. Chem. 2020, 11, 3994–4004. [Google Scholar] [CrossRef]
- Dumeignil, F.; Guehl, M.; Gimbernat, A.; Capron, M.; Ferreira, N.L.; Froidevaux, R.; Girardon, J.-S.; Wojcieszak, R.; Dhulster, P.; Delcroix, D. From Sequential Chemoenzymatic Synthesis to Integrated Hybrid Catalysis: Taking the Best of Both Worlds to Open up the Scope of Possibilities for a Sustainable Future. Catal. Sci. Technol. 2018, 8, 5708–5734. [Google Scholar] [CrossRef]
- Buzatu, A.R.; Frissen, A.E.; van den Broek, L.A.M.; Todea, A.; Motoc, M.; Boeriu, C.G. Chemoenzymatic Synthesis of New Aromatic Esters of Mono- and Oligosaccharides. Processes 2020, 8, 1638. [Google Scholar] [CrossRef]
- Sangiorgio, S.; Pargoletti, E.; Rabuffetti, M.; Robescu, M.S.; Semproli, R.; Ubiali, D.; Cappelletti, G.; Speranza, G. Emulsifying Properties of Sugar-Based Surfactants Prepared by Chemoenzymatic Synthesis. Colloid. Interface Sci. Commun. 2022, 48, 100630. [Google Scholar] [CrossRef]
- Heuson, E.; Froidevaux, R.; Itabaiana, I.; Wojcieszak, R.; Capron, M.; Dumeignil, F. Optimisation of Catalysts Coupling in Multi-Catalytic Hybrid Materials: Perspectives for the next Revolution in Catalysis. Green Chem. 2021, 23, 1942–1954. [Google Scholar] [CrossRef]
- Szelwicka, A.; Zawadzki, P.; Sitko, M.; Boncel, S.; Czardybon, W.; Chrobok, A. Continuous Flow Chemo-Enzymatic Baeyer–Villiger Oxidation with Superactive and Extra-Stable Enzyme/Carbon Nanotube Catalyst: An Efficient Upgrade from Batch to Flow. Org. Process Res. Dev. 2019, 23, 1386–1395. [Google Scholar] [CrossRef]
- Ji, Q.; Wang, B.; Tan, J.; Zhu, L.; Li, L. Immobilized Multienzymatic Systems for Catalysis of Cascade Reactions. Process Biochem. 2016, 51, 1193–1203. [Google Scholar] [CrossRef]
- Ruela, H.S.; Sutili, F.K.; Leal, I.C.R.; Carvalho, N.M.F.; Miranda, L.S.M.; de Souza, R.O.M.A. Lipase-Catalyzed Synthesis of Secondary Glucose Esters under Continuous Flow Conditions. Eur. J. Lipid Sci. Technol. 2013, 115, 464–467. [Google Scholar] [CrossRef]
- Du, L.-H.; Luo, X.-P. Lipase-Catalyzed Regioselective Acylation of Sugar in Microreactors. RSC Adv. 2012, 2, 2663–2665. [Google Scholar] [CrossRef]
- Kim, S.; Ga, S.; Bae, H.; Sluyter, R.; Konstantinov, K.; Kumar Shrestha, L.; Ho Kim, Y.; Ho Kim, J.; Ariga, K. Multidisciplinary Approaches for Enzyme Biocatalysis in Pharmaceuticals: Protein Engineering, Computational Biology, and Nanoarchitectonics. EES Catal. 2024, 2, 14–48. [Google Scholar] [CrossRef]
- Romero-Fernández, M.; Paradisi, F. Protein Immobilization Technology for Flow Biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Farkas, E.; Oláh, M.; Földi, A.; Kóti, J.; Éles, J.; Nagy, J.; Gal, C.A.; Paizs, C.; Hornyánszky, G.; Poppe, L. Chemoenzymatic Dynamic Kinetic Resolution of Amines in Fully Continuous-Flow Mode. Org. Lett. 2018, 20, 8052–8056. [Google Scholar] [CrossRef] [PubMed]
- Brandão, L.M.d.S.; Barbosa, M.S.; Souza, R.L.; Pereira, M.M.; Lima, Á.S.; Soares, C.M.F. Lipase Activation by Molecular Bioimprinting: The Role of Interactions between Fatty Acids and Enzyme Active Site. Biotechnol. Progress. 2021, 37, e3064. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yin, L.; Feng, K.; Zhou, L.; Ma, L.; He, Y.; Wang, L.; Jiang, Y. Lipase Immobilization through the Combination of Bioimprinting and Cross-Linked Protein-Coated Microcrystal Technology for Biodiesel Production. Ind. Eng. Chem. Res. 2016, 55, 11037–11043. [Google Scholar] [CrossRef]
- Carvalho de Melo, J.J.; Passos da Silva, G.L.; Mota, D.A.; de Souza Brandão, L.M.; de Souza, R.L.; Pereira, M.M.; Lima, Á.S.; Soares, C.M.F. Use of Bioprinted Lipases in Microwave-Assisted Esterification Reactions. Catalysts 2023, 13, 299. [Google Scholar] [CrossRef]
- Wiltschi, B.; Cernava, T.; Dennig, A.; Galindo Casas, M.; Geier, M.; Gruber, S.; Haberbauer, M.; Heidinger, P.; Herrero Acero, E.; Kratzer, R.; et al. Enzymes Revolutionize the Bioproduction of Value-Added Compounds: From Enzyme Discovery to Special Applications. Biotechnol. Adv. 2020, 40, 107520. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Bayer, T.; Badenhorst, C.P.S.; Wu, S.; Doerr, M.; Höhne, M.; Bornscheuer, U.T. Recent Trends in Biocatalysis. Chem. Soc. Rev. 2021, 50, 8003–8049. [Google Scholar] [CrossRef] [PubMed]
- Danielli, C.; van Langen, L.; Boes, D.; Asaro, F.; Anselmi, S.; Provenza, F.; Renzi, M.; Gardossi, L. 2,5-Furandicarboxaldehyde as a Bio-Based Crosslinking Agent Replacing Glutaraldehyde for Covalent Enzyme Immobilization. RSC Adv. 2022, 12, 35676–35684. [Google Scholar] [CrossRef]
- Buchholz, P.C.F.; Vogel, C.; Reusch, W.; Pohl, M.; Rother, D.; Spieß, A.C.; Pleiss, J. BioCatNet: A Database System for the Integration of Enzyme Sequences and Biocatalytic Experiments. Chembiochem 2016, 17, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev. MMBR 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Bell, E.L.; Finnigan, W.; France, S.P.; Green, A.P.; Hayes, M.A.; Hepworth, L.J.; Lovelock, S.L.; Niikura, H.; Osuna, S.; Romero, E.; et al. Biocatalysis. Nat. Rev. Methods Primers 2021, 1, 46. [Google Scholar] [CrossRef]
- Almeida, J.M.; Alnoch, R.C.; Souza, E.M.; Mitchell, D.A.; Krieger, N. Metagenomics: Is It a Powerful Tool to Obtain Lipases for Application in Biocatalysis? Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140320. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the Third Wave of Biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Damborsky, J.; Brezovsky, J. Computational Tools for Designing and Engineering Enzymes. Curr. Opin. Chem. Biol. 2014, 19, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Dorn, M.; e Silva, M.B.; Buriol, L.S.; Lamb, L.C. Three-Dimensional Protein Structure Prediction: Methods and Computational Strategies. Comput. Biol. Chem. 2014, 53, 251–276. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Arnold, F.H. Engineering New Catalytic Activities in Enzymes. Nat. Catal. 2020, 3, 203–213. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, V.; Dubey, V.K.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Industrial Applications of Fungal Lipases: A Review. Front. Microbiol. 2023, 14, 1142536. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Sharma, P.K.; Kato, M.; Parson, W.W. Modeling Electrostatic Effects in Proteins. Biochim. Biophys. Acta 2006, 1764, 1647–1676. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.H.; Maiangwa, J.; Nezhad, N.G.; Ali, M.S.M.; Normi, Y.M.; Shariff, F.M.; Rahman, R.N.Z.R.A.; Leow, T.C. Knotting Terminal Ends of Mutant T1 Lipase with Disulfide Bond Improved Structure Rigidity and Stability. Appl. Microbiol. Biotechnol. 2023, 107, 1673–1686. [Google Scholar] [CrossRef]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The Role of Dynamic Conformational Ensembles in Biomolecular Recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Chen, M.; Jin, T.; Chong, W.; Zhang, Z.; Nian, B.; Hu, Y. Computer-Aided Engineering of Lipases Solvent Tolerance Enhanced Their Applications in Sugar Esters Synthesis: State of the Art. Trends Food Sci. Technol. 2024, 144, 104323. [Google Scholar] [CrossRef]
- Reetz, M.T.; Qu, G.; Sun, Z. Engineered Enzymes for the Synthesis of Pharmaceuticals and Other High-Value Products. Nat. Synth. 2024, 3, 19–32. [Google Scholar] [CrossRef]
- Wahab, R.A.; Basri, M.; Rahman, M.B.A.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Chor, L.T. Engineering Catalytic Efficiency of Thermophilic Lipase from Geobacillus Zalihae by Hydrophobic Residue Mutation near the Catalytic Pocket. Adv. Biosci. Biotechnol. 2012, 3, 158–167. [Google Scholar] [CrossRef]
- Kumari, A.; Gupta, R. Phenylalanine to Leucine Point Mutation in Oxyanion Hole Improved Catalytic Efficiency of Lip12 from Yarrowia Lipolytica. Enzym. Microb. Technol. 2013, 53, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Khaleghinejad, S.H.; Motalleb, G.; Karkhane, A.A.; Aminzadeh, S.; Yakhchali, B. Study the Effect of F17S Mutation on the Chimeric Bacillus Thermocatenulatus Lipase. J. Genet. Eng. Biotechnol. 2016, 14, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Ma’ruf, I.F.; Widhiastuty, M.P.; Suharti; Moeis, M.R. Akhmaloka Effect of Mutation at Oxyanion Hole Residu (H110F) on Activity of Lk4 Lipase. Biotechnol. Rep. 2021, 29, e00590. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Fernandes, P. Machine Learning: A Suitable Method for Biocatalysis. Catalysts 2023, 13, 961. [Google Scholar] [CrossRef]
- Ortega-Requena, S.; Montiel, C.; Máximo, F.; Gómez, M.; Murcia, M.D.; Bastida, J. Esters in the Food and Cosmetic Industries: An Overview of the Reactors Used in Their Biocatalytic Synthesis. Materials 2024, 17, 268. [Google Scholar] [CrossRef]
- Sarmah, N.; Mehtab, V.; Bugata, L.S.P.; Tardio, J.; Bhargava, S.; Parthasarathy, R.; Chenna, S. Machine Learning Aided Experimental Approach for Evaluating the Growth Kinetics of Candida Antarctica for Lipase Production. Bioresour. Technol. 2022, 352, 127087. [Google Scholar] [CrossRef]
- Mazurenko, S.; Prokop, Z.; Damborsky, J. Machine Learning in Enzyme Engineering. ACS Catal. 2020, 10, 1210–1223. [Google Scholar] [CrossRef]
- Pleiss, J. Standardized Data, Scalable Documentation, Sustainable Storage—EnzymeML As A Basis For FAIR Data Management In Biocatalysis. ChemCatChem 2021, 13, 3909–3913. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Bucholska, J.; Starowicz, P.; Czyrko, E. Internet Databases of the Properties, Enzymatic Reactions, and Metabolism of Small Molecules—Search Options and Applications in Food Science. Int. J. Mol. Sci. 2016, 17, 2039. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.R.; Alnoch, R.C.; de Almeida, J.M.; dos Santos, L.A.; Andretta, A.T.; Ropaín, R.d.P.C.; de Souza, E.M.; Mitchell, D.A.; Krieger, N. Key Mutation Sites for Improvement of the Enantioselectivity of Lipases through Protein Engineering. Biochem. Eng. J. 2021, 172, 108047. [Google Scholar] [CrossRef]
- Yoshida, K.; Kawai, S.; Fujitani, M.; Koikeda, S.; Kato, R.; Ema, T. Enhancement of Protein Thermostability by Three Consecutive Mutations Using Loop-Walking Method and Machine Learning. Sci. Rep. 2021, 11, 11883. [Google Scholar] [CrossRef]
- Finnigan, W.; Hepworth, L.J.; Flitsch, S.L.; Turner, N.J. RetroBioCat as a Computer-Aided Synthesis Planning Tool for Biocatalytic Reactions and Cascades. Nat. Catal. 2021, 4, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Probst, D.; Manica, M.; Nana Teukam, Y.G.; Castrogiovanni, A.; Paratore, F.; Laino, T. Biocatalysed Synthesis Planning Using Data-Driven Learning. Nat. Commun. 2022, 13, 964. [Google Scholar] [CrossRef] [PubMed]

| Acyl Acceptor | Enzyme | Acyl Donor | Product | Solvent | Ref. |
|---|---|---|---|---|---|
| d-Allose | Novozym435® (Candida antarctica immobilized lipase B) | Vinyl caprylate | 1,6-diacyl-d-psicofuranoses | Acetone, acetonitrile, or tetrahydrofuran (THF) | [54] |
| d-Arabitol | Novozym435® | Lauric acid | 1,5-dilauryl-d-arabitol | Reactive natural deep eutetic solvent (R-NADES) ChCl:d-arabitol | [55] |
| d-Fructose | Immobilized Rhizomucor miehei lipase | Oleic acid | Fructose oleate * | 2-methyl-2-butanol (2M2B) | [56] |
| Novozym435® | Lauric acid | 6-O-lauroyl-d-fructofuranose | Ethyl-methyl ketone | [57] | |
| Novozym435® | Myristic acid | d-fructosyl myristate * | Tert-butanol (tert-BuOH):pyridine (11:9, v/v) | [58] | |
| d-Fructose | Immobilized Rhizomucor miehei lipase | Myristic acid | Fructose myristate * | Solvent-free | [59] |
| d-Galactose | Immobilized Rhizomucor miehei lipase | Oleic acid | Galactose oleate * | [Bmim][BF4]/dimethyl sulfoxide (DMSO) 1-Butyl-3-methylimidazolium tetrafluoroborate | [60] |
| d-Glucose | Aspergillus niger lipase | Lauric acid | 6-O-lauroyl-d-glucopyranose | 2M2B | [61] |
| Aspergillus oryzae lipase | |||||
| Lipozyme TL IM® (Thermomyces lanuginosus immobilized) | |||||
| Novozym435® | |||||
| Novozym435® | Myristic acid | Glucosylmyristate * | 2M2B | [62] | |
| d-Maltose | Novozym435® | Ethyl Butanoate | 6-O-butyrylmaltose | tert-BuOH | [63] |
| Lactose | Candida antarctica lipase (Novozym435® and immobilized on Immobead) | Lauric acid | Lactose monolaurate * | Acetone | [64] |
| Lactose | Free lipase (MAK Wood) | Lauric acid | Lactose monolaurate * | Acetone | [64] |
| L-Rhamnose | Free Pseudomonas stutzeri lipase | Lauric acid | 4-O-lauroyl-rhamnose | Anhydrous THF | [65] |
| d-Maltotriose | Thermomyces lanuginosus lipase (immobilized on celite and granulated with silica (Lipozyme TL IM®)) | Vinyl laurate | 6-O-lauroyl-maltotriose | 2M2B/DMSO (5 or 20% v/v) | [66] |
| Solvent | Novozym435® Activity at 45 °C (µmol·min−1·g−1) | Glucose Solubility at 45 °C after 24 h Incubation (mM) | LogP | Boiling Point (°C) | References |
|---|---|---|---|---|---|
| N,N-dimethylformamide (DMF) | 0 | 12 | −1.0 | 153 | [81,84,85] |
| DMSO | 0 | 29 | −1.35 | 189 | [81,84,85] |
| n-hexane | 0 | 0 | 3.9 | 69 | [81,84,86] |
| THF | 1.6 | 2.1 | 0.46 | 65 | [81,84,86] |
| tert-BuOH | 3.7 | 12 | 0.35 | 82 | [81,84,86] |
| 2M2B | 3.6 | 10 | 0.89 | 102 | [81,84,86] |
| Pyridine | 0 | 134 | 0.65 | 115 | [81,84,87] |
| Saccharide (Carbohydrate) | Acyl Donor | Enzyme | Solvents (v/v) | Time Temperature Pressure | Conversion of Fatty Acyl Donor or Yield * | Ref. |
|---|---|---|---|---|---|---|
| d-Allose | Vinyl esters | Novozym435® | Acetonitrile | 24 h 45 °C atm | 83% * | [54] |
| d-Arabitol | Lauric acid | Novozym435® | R-NADES (reactive natural deep eutetic solvent) ChCl:d-Arabitol | 24 h 70 °C atm | 95% * | [55] |
| d-Fructose | Lauric acid | Novozym435® | [Bmim][TFO] */2M2B (3:2) * 1-Butyl-3-methylimidazolium/trifluoromethanesulfonate | 12 h 50 °C atm | 85% * | [57] |
| Oleic acid | Immobilized Candida rugosa lipase | Solvent-free | 48 h 60 °C atm | 80% * | [92] | |
| Oleic acid | Immobilized Rhizomucor miehei lipase | Solvent-free | 144 h 65 °C atm | 92% | [93] | |
| Palmitic acid | Novozym435® | 2M2B | 72 h 40 °C atm | 78% * | [94] | |
| d-Galactose | Oleic acid | Immobilized Candida rugosa lipase | DMSO/IL[Bmim][BF4] * (1:20) | 2 h 60 °C atm | 87% | [60] |
| d-Glucose | Lauric acid | Novozym435® | 2MeTHF (2-methyltetrahydrofuran) | 72 h 75 °C atm | 48% * | [95] |
| 2MeTHF3one (2-methyltetrahydrofuran-3-one) | 79% * | |||||
| Ethyl laurate | Supported Aspergillus niger lipase | 2M2B/2MeTHF3one (4:1) | 48 h 56 °C atm | 49% * | [96] | |
| Vinyl laurate | 80% * | |||||
| Lauric acid | Novozym435® | DMSO/tert-BuOH (4:1) | 24 h 55 °C atm | 77% | [97] | |
| Palmitic acid | Acetone saturated with supercritical CO2 in continuous reactor | 4 h 50 °C 65 bar | >20% | [91] | ||
| Vinyl palmitate | Acetonitrile | 72 h 45 °C atm | 100% * | [98] | ||
| d-Maltose | Lauric acid | Novozym435® | Acetone/n-hexane (3:2) | 72 h 50 °C atm | 69% | [82] |
| d-Mannose | Capric acid | Immobilized Candida rugosa lipase | Acetone | 48 h 50 °C atm | 62% | [99] |
| Lauric acid | Novozym435® | n-hexane/acetone (1:1) | 72 h 50 °C atm | 25% * | [100] | |
| Vinyl myristate | Novozym435® | [Bmpyrr] */[TFO] * 1-butyl-1-methylpyrrolidinium | 24 h 60 °C atm | 71% * | [101] | |
| d-Xylose | Vinyl laurate | Novozym435® | 2M2B | 4 h 60 °C atm | 25% | [102] |
| Hexanoic acid | Novozym435® | DMSO/Acetone (1:10) | 24 h 60 °C Atm | 64% | [103] |
| Reaction System | Advantages | Limits | References |
|---|---|---|---|
| Solvent |
|
| [69,82,83,88,118] |
| Co-solvent |
|
| [105,119] |
| Solvent-free |
|
| [90,109] |
| Supercritical CO2/solvent |
|
| [91,112] |
| Ionic liquids |
|
| [60,94,120] |
| DES |
|
| [113,114] |
| Abbreviation | Full Name | |
|---|---|---|
| Organic solvent | 2M2B DMSO | 2-methyl-2-butanol Dimethyl sulfoxide |
| DMF | N,N-dimethylformamide | |
| THF | Tetrahydrofuran | |
| 2MeTHF | 2-methyltetrahydrofuran | |
| 2MeTHF3one | 2-methyltetrahydrofuran-3-one | |
| R-NADES | Reactive natural deep eutetic solvent | |
| ChCl | Choline chloride | |
| tert-BuOH | Tert-butanol | |
| [Bmim][BF4] | 1-Butyl-3-methylimidazolium/tetrafluoroborate | |
| [Bmim][TFO] | 1-Butyl-3-methylimidazolium/ trifluoromethanesulfonate | |
| [Bmpyrr] | 1-butyl-1-methylpyrrolidinium | |
| IL | Ionic liquid | |
| DES | Deep eutetic solvent | |
| Lipase | CalB | Lipase B from Candida antarctica |
| TLL | Lipase from Thermomyces lanuginosus | |
| Biocatalyst | Novozym435® | Lipase B from Candida antarctica produced by Novozymes |
| Lipozyme TL IM® | Recombinant TLL immobilized on silica |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spalletta, A.; Joly, N.; Martin, P. Latest Trends in Lipase-Catalyzed Synthesis of Ester Carbohydrate Surfactants: From Key Parameters to Opportunities and Future Development. Int. J. Mol. Sci. 2024, 25, 3727. https://doi.org/10.3390/ijms25073727
Spalletta A, Joly N, Martin P. Latest Trends in Lipase-Catalyzed Synthesis of Ester Carbohydrate Surfactants: From Key Parameters to Opportunities and Future Development. International Journal of Molecular Sciences. 2024; 25(7):3727. https://doi.org/10.3390/ijms25073727
Chicago/Turabian StyleSpalletta, Alexis, Nicolas Joly, and Patrick Martin. 2024. "Latest Trends in Lipase-Catalyzed Synthesis of Ester Carbohydrate Surfactants: From Key Parameters to Opportunities and Future Development" International Journal of Molecular Sciences 25, no. 7: 3727. https://doi.org/10.3390/ijms25073727







