Deep Learning Approach for Differentiating Etiologies of Pediatric Retinal Hemorrhages: A Multicenter Study
Abstract
:1. Introduction
2. Results
3. Discussion
4. Methods and Materials
4.1. Study Population
4.2. Annotation
4.3. Data Preprocessing
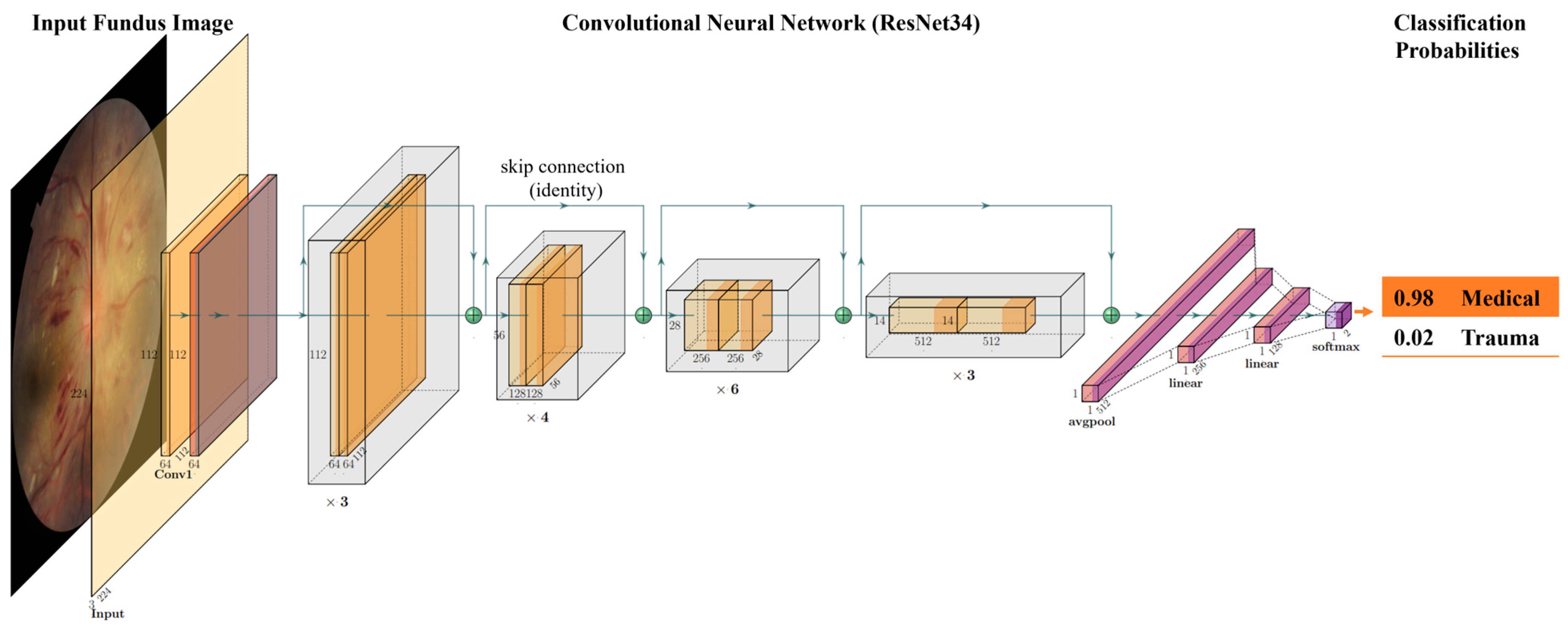
4.4. Algorithm Development
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thau, A.; Saffren, B.; Zakrzewski, H.; Anderst, J.D.; Carpenter, S.L.; Levin, A. Retinal hemorrhage and bleeding disorders in children: A review. Child Abus. Negl. 2021, 112, 104901. [Google Scholar] [CrossRef] [PubMed]
- Di Fazio, N.; Delogu, G.; Morena, D.; Cipolloni, L.; Scopetti, M.; Mazzilli, S.; Frati, P.; Fineschi, V. New Insights into the Diagnosis and Age Determination of Retinal Hemorrhages from Abusive Head Trauma: A Systematic Review. Diagnostics 2023, 13, 1722. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.V. Retinal hemorrhage: Science versus speculation. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2016, 20, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Yiu, G.; Mahmoud, T.H. Subretinal Hemorrhage. Dev. Ophthalmol. 2014, 54, 213–222. [Google Scholar] [CrossRef]
- Forbes, B.J.; Rubin, S.E.; Margolin, E.; Levin, A.V. Evaluation and management of retinal hemorrhages in infants with and without abusive head trauma. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2010, 14, 267–273. [Google Scholar] [CrossRef]
- Watts, P.; Maguire, S.; Kwok, T.; Talabani, B.; Mann, M.; Wiener, J.; Lawson, Z.; Kemp, A. Newborn retinal hemorrhages: A systematic review. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2013, 17, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Moshfeghi, D.M. Terson Syndrome in a Healthy Term Infant: Delivery-Associated Retinopathy and Intracranial Hemorrhage. Ophthalmic Surg. Lasers Imaging Retin. 2018, 49, e154–e156. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, K.; Stoessel, K.; Leventhal, J.M.; Ogle, E.; Teague, B.; Lavietes, S.; Banyas, B.; Allen, K.; Dziura, J.; Duncan, C. Characteristics That Distinguish Accidental from Abusive Injury in Hospitalized Young Children with Head Trauma. Pediatrics 2004, 114, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Song, H.H.; Thoreson, W.B.; Dong, P.; Shokrollahi, Y.; Gu, L.; Suh, D.W. Exploring the Vitreoretinal Interface: A Key Instigator of Unique Retinal Hemorrhage Patterns in Pediatric Head Trauma. Korean J. Ophthalmol. 2022, 36, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Morad, Y.; Wygnansky-Jaffe, T.; Levin, A.V. Retinal haemorrhage in abusive head trauma. Clin. Exp. Ophthalmol. 2010, 38, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Togioka, B.M.; Arnold, M.A.; Bathurst, M.A.; Ziegfeld, S.M.; Nabaweesi, R.; Colombani, P.M.; Chang, D.C.; Abdullah, F. Retinal Hemorrhages and Shaken Baby Syndrome: An Evidence-Based Review. J. Emerg. Med. 2008, 37, 98–106. [Google Scholar] [CrossRef]
- Falavarjani, K.G.; Parvaresh, M.M.; Shahraki, K.; Nekoozadeh, S.; Amirfarhangi, A. Central retinal artery occlusion in Crohn disease. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2012, 16, 392–393. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, G.; Chen, W.; Huang, J.; Ying, G.-S.; Forbes, B.J. The natural history of retinal hemorrhage in pediatric head trauma. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2016, 20, 131–135. [Google Scholar] [CrossRef]
- Sengupta, S.; Singh, A.; Leopold, H.A.; Gulati, T.; Lakshminarayanan, V. Ophthalmic diagnosis using deep learning with fundus images—A critical review. Artif. Intell. Med. 2019, 102, 101758. [Google Scholar] [CrossRef]
- Balyen, L.; Peto, T. Promising Artificial Intelligence-Machine Learning-Deep Learning Algorithms in Ophthalmology. Asia-Pacific J. Ophthalmol. 2019, 8, 264–272. [Google Scholar] [CrossRef]
- Ting, D.S.; Peng, L.; Varadarajan, A.V.; Keane, P.A.; Burlina, P.M.; Chiang, M.F.; Schmetterer, L.; Pasquale, L.R.; Bressler, N.M.; Webster, D.R.; et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog. Retin. Eye Res. 2019, 72, 100759. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef]
- Tsiknakis, N.; Theodoropoulos, D.; Manikis, G.; Ktistakis, E.; Boutsora, O.; Berto, A.; Scarpa, F.; Scarpa, A.; Fotiadis, D.I.; Marias, K. Deep learning for diabetic retinopathy detection and classification based on fundus images: A review. Comput. Biol. Med. 2021, 135, 104599. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-H.; Liu, T.Y.A. Application of Deep Learning to Retinal-Image-Based Oculomics for Evaluation of Systemic Health: A Review. J. Clin. Med. 2022, 12, 152. [Google Scholar] [CrossRef]
- Panda, N.R.; Sahoo, A.K. A Detailed Systematic Review on Retinal Image Segmentation Methods. J. Digit. Imaging 2022, 35, 1250–1270. [Google Scholar] [CrossRef]
- Moradi, M.; Chen, Y.; Du, X.; Seddon, J.M. Deep ensemble learning for automated non-advanced AMD classification using optimized retinal layer segmentation and SD-OCT scans. Comput. Biol. Med. 2023, 154, 106512. [Google Scholar] [CrossRef] [PubMed]
- Prahs, P.; Radeck, V.; Mayer, C.; Cvetkov, Y.; Cvetkova, N.; Helbig, H.; Märker, D. OCT-based deep learning algorithm for the evaluation of treatment indication with anti-vascular endothelial growth factor medications. Graefe’s Arch. Clin. Exp. Ophthalmol. 2017, 256, 91–98. [Google Scholar] [CrossRef]
- Campbell, J.P.; Chiang, M.F.; Chen, J.S.; Moshfeghi, D.M.; Nudleman, E.; Cherwek, H.; Cheung, C.Y.; Singh, P.; Kalpathy-Cramer, J.; Ostmo, S.; et al. Artificial Intelligence for Retinopathy of Prematurity. Ophthalmology 2022, 129, e69–e76. [Google Scholar] [CrossRef]
- Campbell, J.P.; Singh, P.; Redd, T.K.; Brown, J.M.; Shah, P.K.; Subramanian, P.; Rajan, R.; Valikodath, N.; Cole, E.; Ostmo, S.; et al. Applications of Artificial Intelligence for Retinopathy of Prematurity Screening. Pediatrics 2021, 147, e2020016618. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Acharya, U.R.; Ng, E.Y.K.; Ng, K.H.; Suri, J.S. Algorithms for the Automated Detection of Diabetic Retinopathy Using Digital Fundus Images: A Review. J. Med. Syst. 2010, 36, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, H.; She, C.; Feng, J.; Liu, X.; Hu, X.; Chen, L.; Tao, Y. Artificial intelligence promotes the diagnosis and screening of diabetic retinopathy. Front. Endocrinol. 2022, 13, 946915. [Google Scholar] [CrossRef]
- Keenan, T.D.; Dharssi, S.; Peng, Y.; Chen, Q.; Agrón, E.; Wong, W.T.; Lu, Z.; Chew, E.Y. A Deep Learning Approach for Automated Detection of Geographic Atrophy from Color Fundus Photographs. Ophthalmology 2019, 126, 1533–1540. [Google Scholar] [CrossRef]
- Yan, Q.; Weeks, D.E.; Xin, H.; Swaroop, A.; Chew, E.Y.; Huang, H.; Ding, Y.; Chen, W. Deep-learning-based prediction of late age-related macular degeneration progression. Nat. Mach. Intell. 2020, 2, 141–150. [Google Scholar] [CrossRef]
- Lee, J.; Wanyan, T.; Chen, Q.; Keenan, T.D.L.; Glicksberg, B.S.; Chew, E.Y.; Lu, Z.; Wang, F.; Peng, Y. Predicting Age-related Macular Degeneration Progression with Longitudinal Fundus Images Using Deep Learning. Mach. Learn Med. Imaging 2022, 13583, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Morano, J.; Hervella, S.; Rouco, J.; Novo, J.; Fernández-Vigo, J.I.; Ortega, M. Weakly-supervised detection of AMD-related lesions in color fundus images using explainable deep learning. Comput. Methods Programs Biomed. 2023, 229, 107296. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Su, Y.; Lin, F.; Li, Z.; Song, Y.; Nie, S.; Xu, J.; Chen, L.; Chen, S.; Li, H.; et al. A deep-learning system predicts glaucoma incidence and progression using retinal photographs. J. Clin. Investig. 2022, 132, e157968. [Google Scholar] [CrossRef]
- Coan, L.J.; Williams, B.M.; Adithya, V.K.; Upadhyaya, S.; Alkafri, A.; Czanner, S.; Venkatesh, R.; Willoughby, C.E.; Kavitha, S.; Czanner, G. Automatic detection of glaucoma via fundus imaging and artificial intelligence: A review. Surv. Ophthalmol. 2022, 68, 17–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tang, L.; Xia, M.; Cao, G. The application of artificial intelligence in glaucoma diagnosis and prediction. Front. Cell Dev. Biol. 2023, 11, 1173094. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Lim, J.S.; Foo, L.L.; Ng, W.Y.Y.; Yip, M.; Lim, G.Y.S.; Wong, M.H.Y.; Fong, A.; Rosman, M.; Mehta, J.S.; et al. Application of artificial intelligence in cataract management: Current and future directions. Eye Vis. 2022, 9, 1–11. [Google Scholar] [CrossRef]
- Park, Y.-J.; Bae, J.H.; Shin, M.H.; Hyun, S.H.; Cho, Y.S.; Choe, Y.S.; Choi, J.Y.; Lee, K.-H.; Kim, B.-T.; Moon, S.H. Development of Predictive Models in Patients with Epiphora Using Lacrimal Scintigraphy and Machine Learning. Nucl. Med. Mol. Imaging 2019, 53, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Kuo, B.-I.; Chang, W.-Y.; Liao, T.-S.; Liu, F.-Y.; Liu, H.-Y.; Chu, H.-S.; Chen, W.-L.; Hu, F.-R.; Yen, J.-Y.; Wang, I.-J. Keratoconus Screening Based on Deep Learning Approach of Corneal Topography. Transl. Vis. Sci. Technol. 2020, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Kim, Y.; Shin, K.Y.; Han, S.H.; Oh, S.Y.; Chung, T.-Y.; Park, K.-A.; Lim, D.H. Deep Learning–Based Prediction of Refractive Error Using Photorefraction Images Captured by a Smartphone: Model Development and Validation Study. JMIR Public Heal. Surveill. 2020, 8, e16225. [Google Scholar] [CrossRef]
- Dong, L.; He, W.; Zhang, R.; Ge, Z.; Wang, Y.X.; Zhou, J.; Xu, J.; Shao, L.; Wang, Q.; Yan, Y.; et al. Artificial Intelligence for Screening of Multiple Retinal and Optic Nerve Diseases. JAMA Netw. Open 2022, 5, e229960. [Google Scholar] [CrossRef] [PubMed]
- Milea, D.; Najjar, R.P.; Jiang, Z.; Ting, D.; Vasseneix, C.; Xu, X.; Fard, M.A.; Fonseca, P.; Vanikieti, K.; Lagrèze, W.A.; et al. Artificial Intelligence to Detect Papilledema from Ocular Fundus Photographs. N. Engl. J. Med. 2020, 382, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Charoenlarpnopparut, C.; Mahapakulchai, S. Deep learning-based hemorrhage detection for diabetic retinopathy screening. Sci. Rep. 2023, 13, 1479. [Google Scholar] [CrossRef] [PubMed]
- Skouta, A.; Elmoufidi, A.; Jai-Andaloussi, S.; Ouchetto, O. Hemorrhage semantic segmentation in fundus images for the diagnosis of diabetic retinopathy by using a convolutional neural network. J. Big Data 2022, 9, 78. [Google Scholar] [CrossRef]
- Mao, J.; Luo, Y.; Chen, K.; Lao, J.; Chen, L.; Shao, Y.; Zhang, C.; Sun, M.; Shen, L. New grading criterion for retinal haemorrhages in term newborns based on deep convolutional neural networks. Clin. Exp. Ophthalmol. 2020, 48, 220–229. [Google Scholar] [CrossRef]
- Galdran, A.; Anjos, A.; Dolz, J.; Chakor, H.; Lombaert, H.; Ayed, I.B. State-of-the-art retinal vessel segmentation with minimalistic models. Sci. Rep. 2022, 12, 6174. [Google Scholar] [CrossRef]
- Balasubramanian, V. Statistical Inference, Occam’s Razor, and Statistical Mechanics on the Space of Probability Distributions. Neural Comput. 1997, 9, 349–368. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Devries, T.; Taylor, G.W. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual attention network for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3156–3164. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Vasu, P.K.A.; Gabriel, J.; Zhu, J.; Tuzel, O.; Ranjan, A. FastViT: A Fast Hybrid Vision Transformer using Structural Reparameterization. arXiv 2023, arXiv:2303.14189. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the NeurIPS, Vancouver, BC, Canada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- PyTorch Image Models, 2019, 10.5281/zenodo. 4414861. GitHub Repository. Available online: https://github.com/rwightman/pytorch-image-models (accessed on 1 September 2023).
- Steiner, A.; Kolesnikov, A.; Zhai, X.; Wightman, R.; Uszkoreit, J.; Beyer, L. How to train your vit? data, augmentation, and regularization in vision transformers. arXiv 2021, arXiv:2106.10270. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Bergstra, J.; Yamins, D.; Cox, D.D. Making a Science of Model Search: Hyperparameter Optimization in Hundreds of Dimensions for Vision Architectures. In Proceedings of the 30th International Conference on Machine Learning (ICML 2013), Atlanta, GA, USA, 16–21 June 2013; pp. I-115–I-123. [Google Scholar]
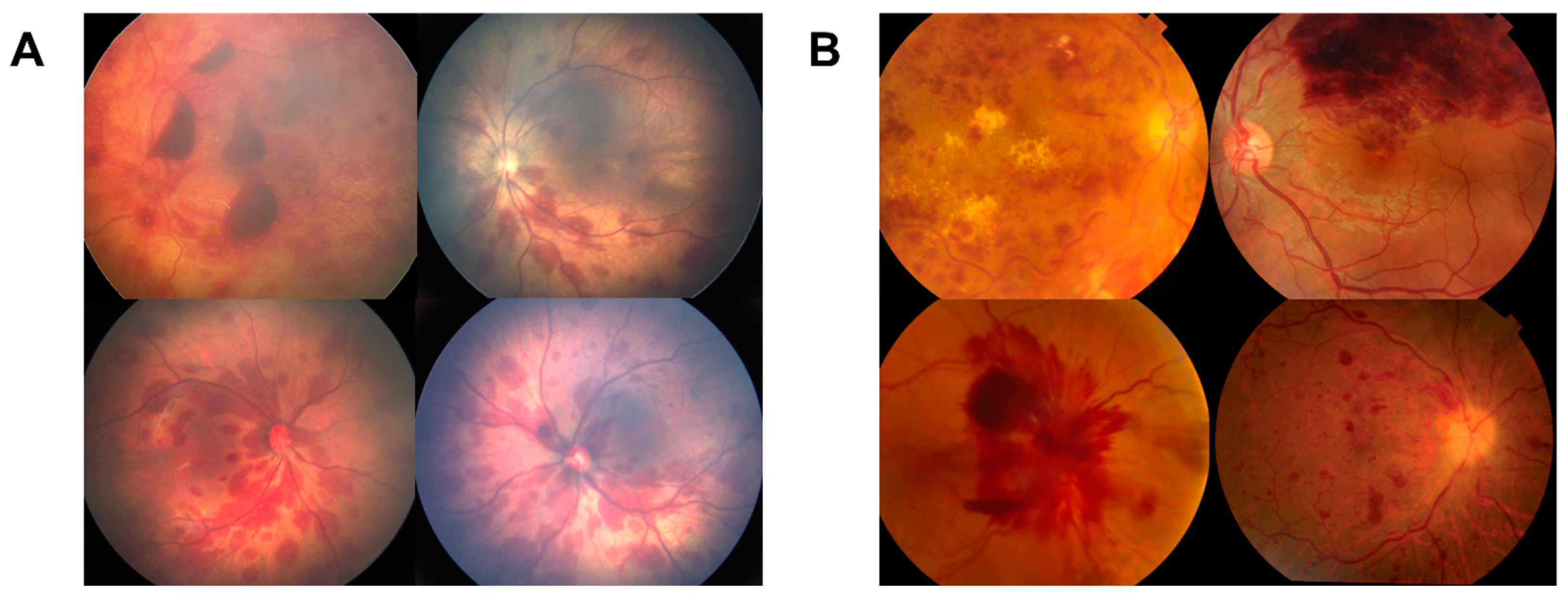
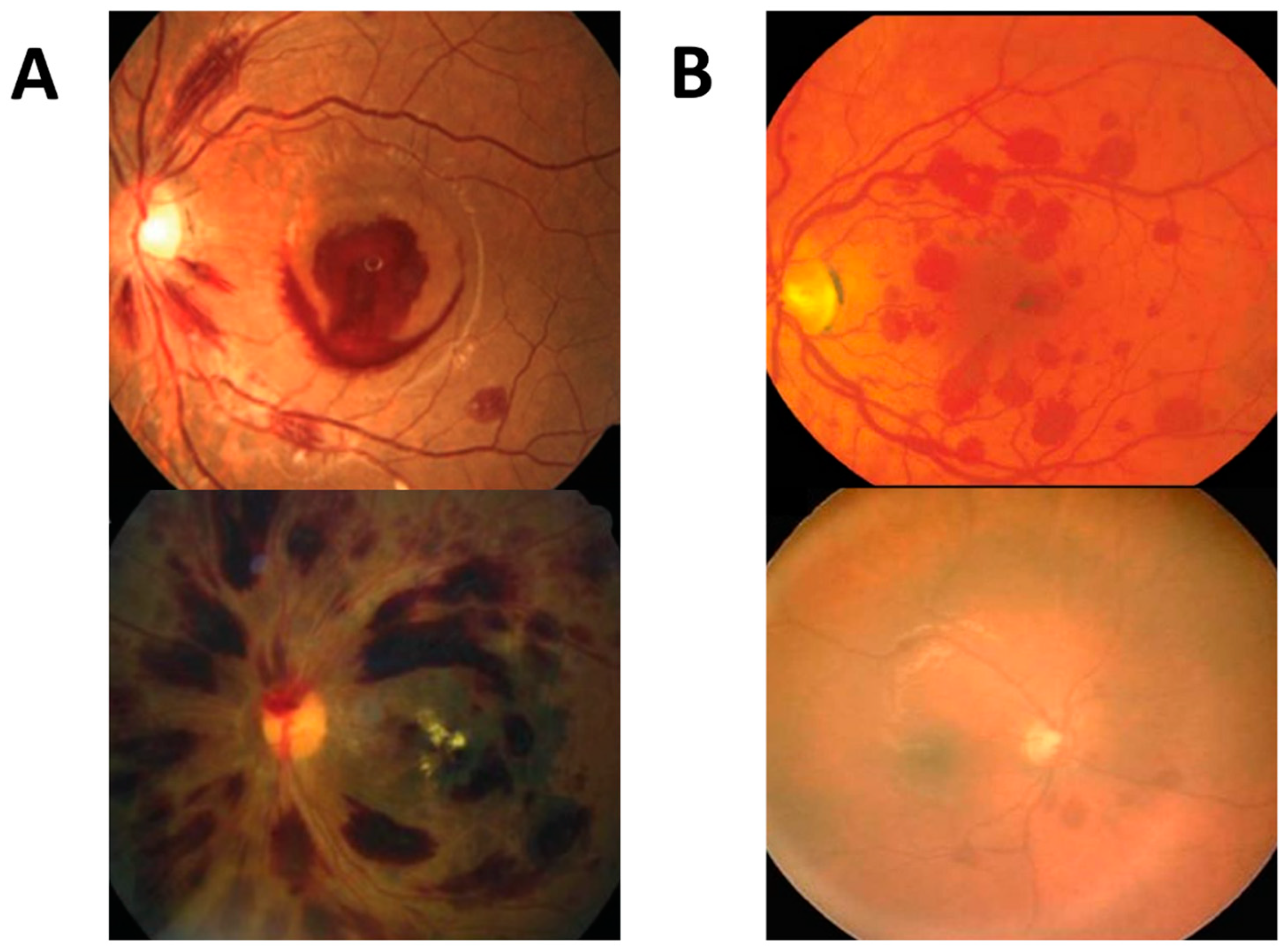

| Diagnosis | All (n = 599) | Training (n = 343) | Validation (n = 127) | Test (n = 127) |
|---|---|---|---|---|
| Medical | 298 | 172 | 61 | 65 |
| Retinal Vascular Disease | 102 | 54 | 20 | 27 |
| Leukemia | 89 | 46 | 16 | 17 |
| Papilledema | 19 | 12 | 4 | 3 |
| Coagulopathy | 88 | 60 | 22 | 18 |
| Trauma | 299 | 171 | 66 | 62 |
| Accidental Trauma | 18 | 11 | 3 | 4 |
| Birth Trauma | 118 | 67 | 25 | 26 |
| Abusive Head Trauma | 163 | 93 | 38 | 32 |
| Model | Accuracy | AUC | Specificity | Sensitivity | PPV | NPV |
|---|---|---|---|---|---|---|
| ResNet18 | 88.98% | 0.9506 | 83.08% | 96.77% | 84.29% | 94.74% |
| ResNet34 | 86.61% | 0.9437 | 87.69% | 87.10% | 86.89% | 86.36% |
| ResNet50 | 87.40% | 0.9467 | 84.62% | 91.94% | 84.85% | 90.16% |
| ResNet101 | 89.76% | 0.9449 | 90.77% | 90.32% | 90.16% | 89.39% |
| ResNet152 | 88.19% | 0.9365 | 84.62% | 93.55% | 85.07% | 91.67% |
| ResAttNet56 | 87.40% | 0.9400 | 83.08% | 93.55% | 83.82% | 91.53% |
| ViT-Small | 79.53% | 0.8945 | 78.46% | 82.26% | 78.12% | 80.95% |
| FastViT-SA12 | 90.55% | 0.9628 | 96.92% | 85.48% | 96.30% | 86.30% |
| FastViT-SA24 | 88.19% | 0.9462 | 87.69% | 90.32% | 87.30% | 89.06% |
| Model | Number of Layers | Trainable Parameters (in Millions) |
|---|---|---|
| ResNet18 | 18 | 11.4 |
| ResNet34 | 32 | 21.5 |
| ResNet50 | 50 | 24.0 |
| ResNet101 | 101 | 43.1 |
| ResNet152 | 152 | 58.7 |
| ResAttNet56 | 56 | 29.8 |
| ViT-Small | 12 | 22.5 |
| FastVit-SA12 | 12 | 10.5 |
| FastVit-SA24 | 24 | 20.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khosravi, P.; Huck, N.A.; Shahraki, K.; Hunter, S.C.; Danza, C.N.; Kim, S.Y.; Forbes, B.J.; Dai, S.; Levin, A.V.; Binenbaum, G.; et al. Deep Learning Approach for Differentiating Etiologies of Pediatric Retinal Hemorrhages: A Multicenter Study. Int. J. Mol. Sci. 2023, 24, 15105. https://doi.org/10.3390/ijms242015105
Khosravi P, Huck NA, Shahraki K, Hunter SC, Danza CN, Kim SY, Forbes BJ, Dai S, Levin AV, Binenbaum G, et al. Deep Learning Approach for Differentiating Etiologies of Pediatric Retinal Hemorrhages: A Multicenter Study. International Journal of Molecular Sciences. 2023; 24(20):15105. https://doi.org/10.3390/ijms242015105
Chicago/Turabian StyleKhosravi, Pooya, Nolan A. Huck, Kourosh Shahraki, Stephen C. Hunter, Clifford Neil Danza, So Young Kim, Brian J. Forbes, Shuan Dai, Alex V. Levin, Gil Binenbaum, and et al. 2023. "Deep Learning Approach for Differentiating Etiologies of Pediatric Retinal Hemorrhages: A Multicenter Study" International Journal of Molecular Sciences 24, no. 20: 15105. https://doi.org/10.3390/ijms242015105






