Age-Related microRNA Overexpression in Lafora Disease Male Mice Provides Links between Neuroinflammation and Oxidative Stress
Abstract
:1. Introduction
2. Results and Discussion
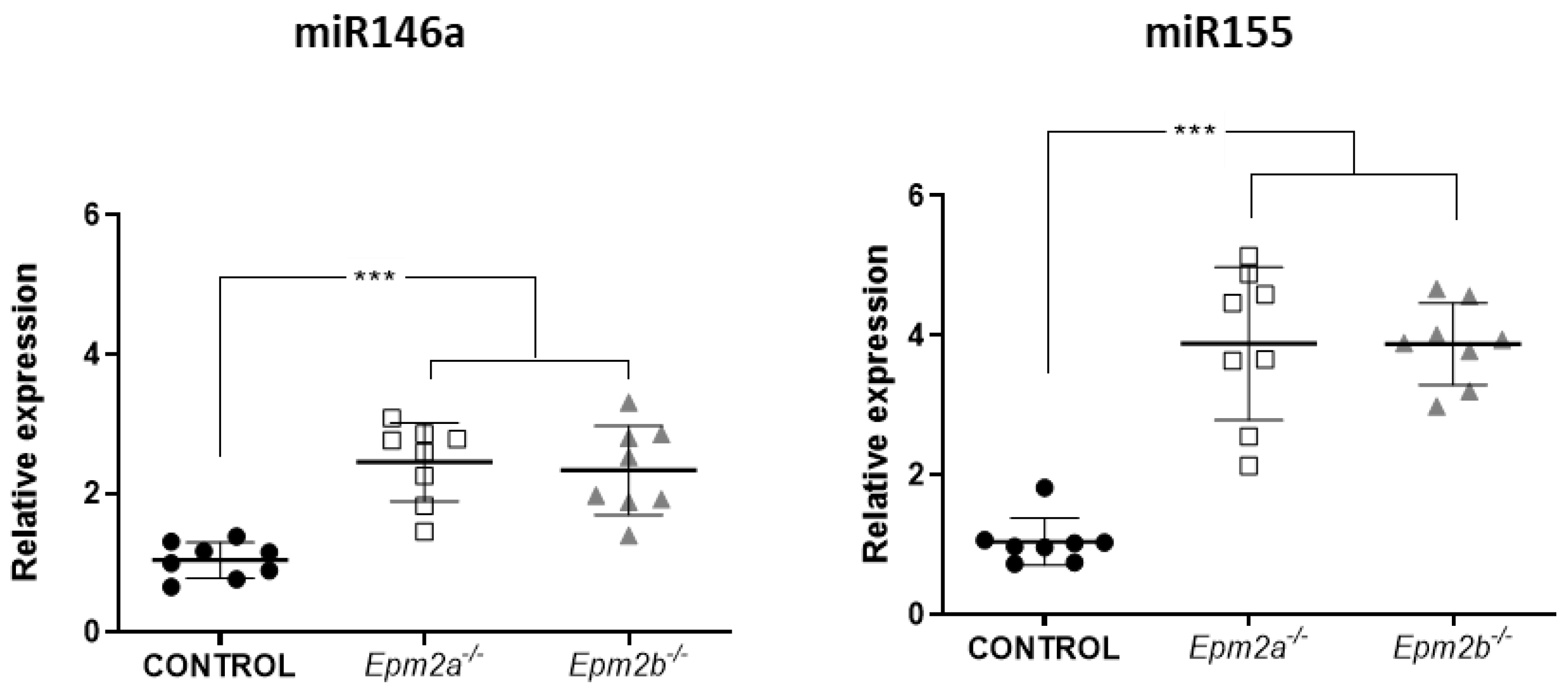
2.1. Differential microRNA Expression in the Brain of 16-Month-Old Epm2a−/− and Epm2b−/− Mice as Compared to Control Mice
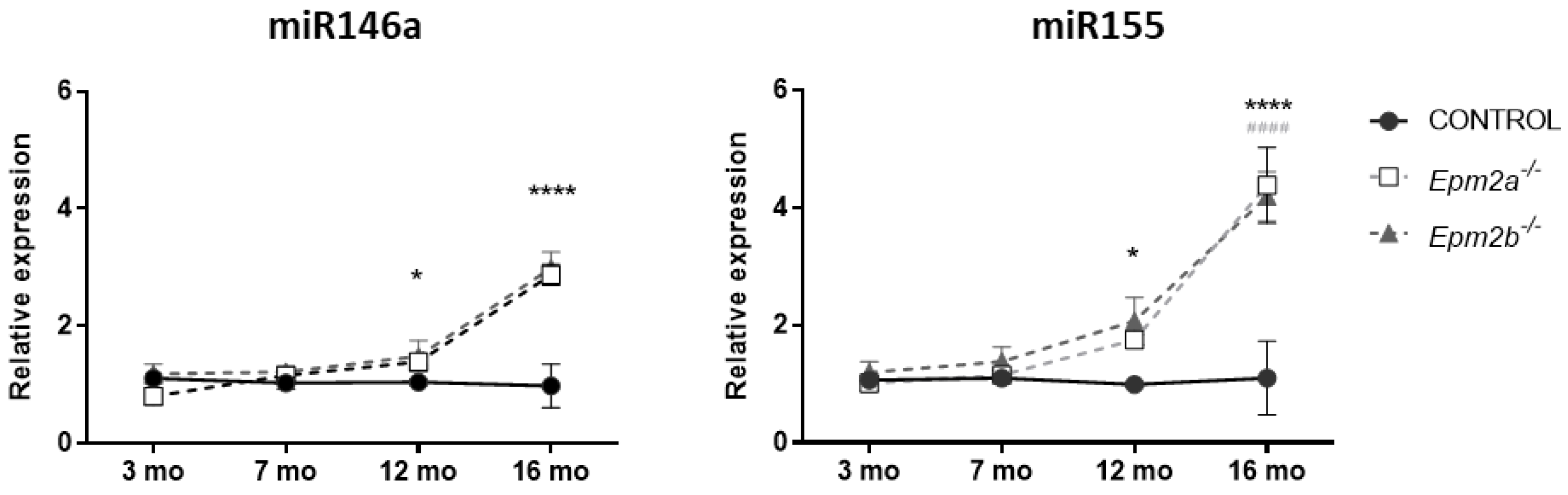
2.2. MicroRNAs miR-155 and miR-146a Are Overexpressed in an Age-Dependent Manner
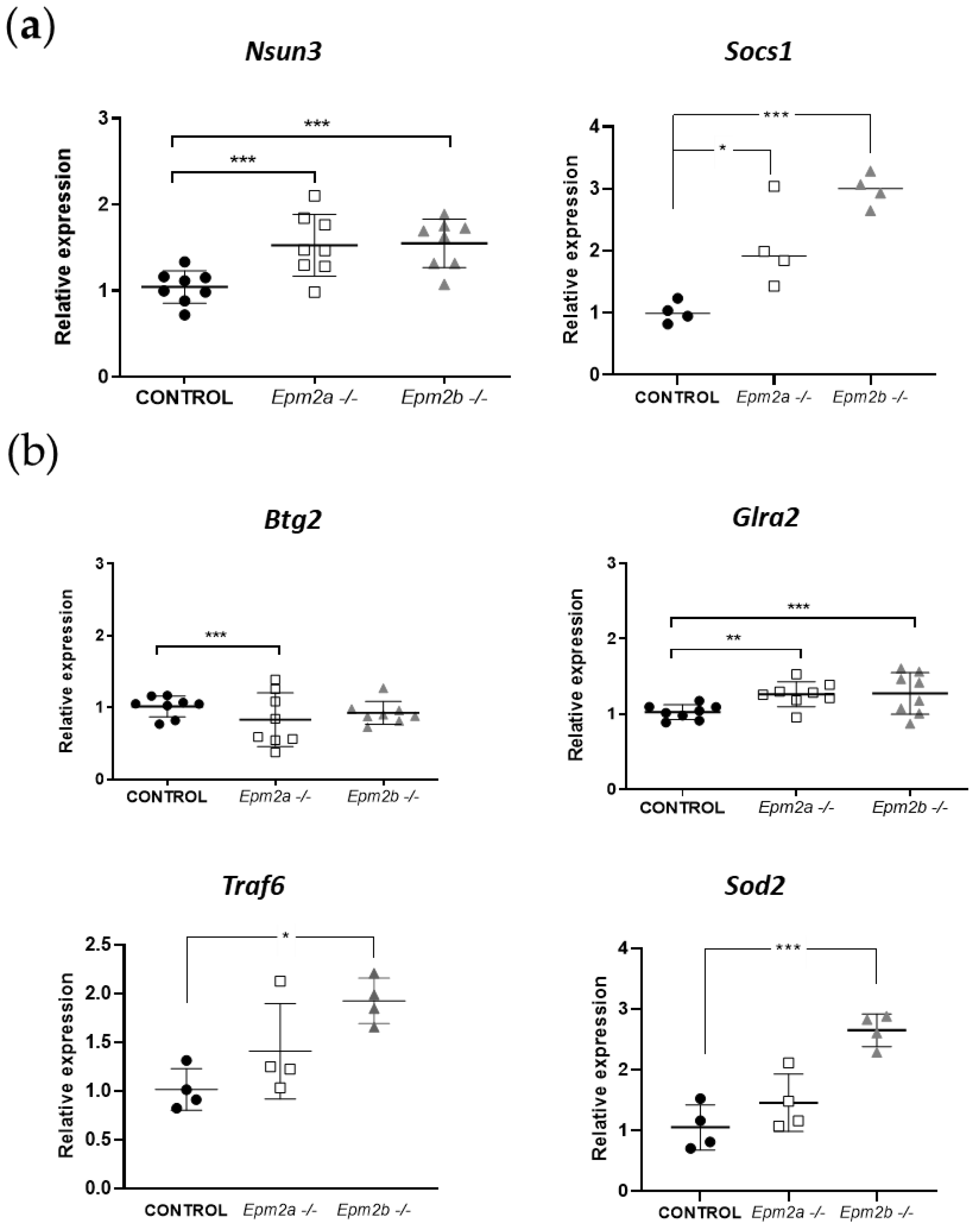
2.3. Analysis of the Expression of Putative Gene Targets of miR-146a and miR-155 in Brain Extracts from Epm2a−/− and Epm2b−/− Mice of 16 Months of Age
3. Materials and Methods
3.1. Animal Care, Mice, and Husbandry
3.2. Whole RNA Extraction from Mouse Brain
3.3. Small RNA-Seq and Data Analysis
3.4. RT-qPCR Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gentry, M.S.; Worby, C.A.; Dixon, J.E. Insights into Lafora Disease: Malin Is an E3 Ubiquitin Ligase That Ubiquitinates and Promotes the Degradation of Laforin. Proc. Natl. Acad. Sci. USA 2005, 102, 8501–8506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worby, C.A.; Gentry, M.S.; Dixon, J.E. Laforin, a Dual Specificity Phosphatase That Dephosphorylates Complex Carbohydrates. J. Biol. Chem. 2006, 281, 30412–30418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pondrelli, F.; Muccioli, L.; Licchetta, L.; Mostacci, B.; Zenesini, C.; Tinuper, P.; Vignatelli, L.; Bisulli, F. Natural History of Lafora Disease: A Prognostic Systematic Review and Individual Participant Data Meta-Analysis. Orphanet J. Rare Dis. 2021, 16, 362. [Google Scholar] [CrossRef] [PubMed]
- Susumu, Y.; Hiroshi, N.; Tasuo, N. Biochemical Studies on Tissues from a Patient with Lafora Disease. Clin. Chim. Acta 1975, 62, 415–423. [Google Scholar] [CrossRef]
- Lafora, G.R.; Glueck, B. Beitrag Zur Histopathologie Der Myoklonischen Epilepsie. Zeitschrift für Die Gesamte Neurologie und Psychiatrie 1911, 6, 1–14. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Turnbull, J.; Wang, W.; Girard, J.M.; Zhao, X.; Skurat, A.V.; Delgado-Escueta, A.V.; Minassian, B.A.; Depaoli-Roach, A.A.; Roach, P.J. Laforin Is a Glycogen Phosphatase, Deficiency of Which Leads to Elevated Phosphorylation of Glycogen in Vivo. Proc. Natl. Acad. Sci. USA 2007, 104, 19262–19266. [Google Scholar] [CrossRef] [Green Version]
- Romá-Mateo, C.; Sanz, P.; Gentry, M.S. Deciphering the Role of Malin in the Lafora Progressive Myoclonus Epilepsy. IUBMB Life 2012, 64, 801–808. [Google Scholar] [CrossRef] [Green Version]
- Romá-Mateo, C.; Aguado, C.; García-Giménez, J.L.; Ibáñez-Cabellos, J.S.; Seco-Cervera, M.; Pallardó, F.V.; Knecht, E.; Sanz, P. Increased Oxidative Stress and Impaired Antioxidant Response in Lafora Disease. Mol. Neurobiol. 2015, 51, 932–946. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Ballester, C.; Berthier, A.; Viana, R.; Sanz, P. Homeostasis of the Astrocytic Glutamate Transporter GLT-1 Is Altered in Mouse Models of Lafora Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2016, 1862, 1074–1083. [Google Scholar] [CrossRef] [Green Version]
- Lahuerta, M.; Gonzalez, D.; Aguado, C.; Fathinajafabadi, A.; García-Giménez, J.L.; Moreno-Estellés, M.; Romá-Mateo, C.; Knecht, E.; Pallardó, F.V.; Sanz, P. Reactive Glia-Derived Neuroinflammation: A Novel Hallmark in Lafora Progressive Myoclonus Epilepsy That Progresses with Age. Mol. Neurobiol. 2020, 57, 1607–1621. [Google Scholar] [CrossRef]
- López-González, I.; Viana, R.; Sanz, P.; Ferrer, I. Inflammation in Lafora Disease: Evolution with Disease Progression in Laforin and Malin Knock-out Mouse Models. Mol. Neurobiol. 2017, 54, 3119–3130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthier, A.; Payá, M.; García-Cabrero, A.M.; Ballester, M.I.; Heredia, M.; Serratosa, J.M.; Sánchez, M.P.; Sanz, P. Pharmacological Interventions to Ameliorate Neuropathological Symptoms in a Mouse Model of Lafora Disease. Mol. Neurobiol. 2015, 53, 1296–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Singh, P.K.; Bhadauriya, P.; Ganesh, S. Lafora Disease E3 Ubiquitin Ligase Malin Is Recruited to the Processing Bodies and Regulates the MicroRNA-Mediated Gene Silencing Process via the Decapping Enzyme Dcp1a. RNA Biol. 2012, 9, 1440–1449. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.-V.; Mastroeni, D.; Audrain, M.; Fanutza, T.; Kim, S.H.; Blitzer, R.D.; Gandy, S.; Dudley, J.T.; Ehrlich, M.E. MiR155 Regulation of Behavior, Neuropathology, and Cortical Transcriptomics in Alzheimer’s Disease. Acta Neuropathol. 2020, 140, 295–315. [Google Scholar] [CrossRef]
- Brennan, G.P.; Henshall, D.C. MicroRNAs as Regulators of Brain Function and Targets for Treatment of Epilepsy. Nat. Rev. Neurol. 2020, 16, 506–519. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. MiR-146 and MiR-155: Two Key Modulators of Immune Response and Tumor Development. Noncoding RNA 2017, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Song, M.; Song, H.; Wang, Y.; Luo, M.; Yin, L. MiR-155 Mediates Inflammatory Injury of Hippocampal Neuronal Cells via the Activation of Microglia. Mol. Med. Rep. 2019, 19, 2627–2635. [Google Scholar] [CrossRef]
- Paramasivam, A.; Meena, A.K.; Venkatapathi, C.; Pitceathly, R.D.S.; Thangaraj, K. Novel Biallelic NSUN3 Variants Cause Early-Onset Mitochondrial Encephalomyopathy and Seizures. J. Mol. Neurosci. 2020, 70, 1962–1965. [Google Scholar] [CrossRef]
- Chen, X.; Wilson, K.A.; Schaefer, N.; de Hayr, L.; Windsor, M.; Scalais, E.; van Rijckevorsel, G.; Stouffs, K.; Villmann, C.; O’Mara, M.L.; et al. Loss, Gain and Altered Function of GlyR A2 Subunit Mutations in Neurodevelopmental Disorders. Front. Mol. Neurosci. 2022, 15, 886729. [Google Scholar] [CrossRef]
- Yildirim, F.; Foddis, M.; Blumenau, S.; Müller, S.; Kajetan, B.; Holtgrewe, M.; Kola, V.; Beule, D.; Sassi, C. Shared and Oppositely Regulated Transcriptomic Signatures in Huntington’s Disease and Brain Ischemia Confirm Known and Unveil Novel Potential Neuroprotective Genes. Neurobiol. Aging 2021, 104, 122.e1–122.e17. [Google Scholar] [CrossRef]
- Semmler, S.; Gagné, M.; Garg, P.; Pickles, S.R.; Baudouin, C.; Hamon-Keromen, E.; Destroismaisons, L.; Khalfallah, Y.; Chaineau, M.; Caron, E.; et al. TNF Receptor–Associated Factor 6 Interacts with ALS-Linked Misfolded Superoxide Dismutase 1 and Promotes Aggregation. J. Biol. Chem. 2020, 295, 3808–3825. [Google Scholar] [CrossRef] [PubMed]
- Zingale, V.D.; Gugliandolo, A.; Mazzon, E. MiR-155: An Important Regulator of Neuroinflammation. Int. J. Mol. Sci. 2021, 23, 90. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs Activate Gene Transcription Epigenetically as an Enhancer Trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, Y.; Guo, J.; Cui, L.; Yang, L.; Li, Y.; Mou, Y.; Jia, C.; Zhang, L.; Song, X. MiR-146a Enhances Regulatory T-cell Differentiation and Function in Allergic Rhinitis by Targeting STAT5b. Allergy 2022, 77, 550–558. [Google Scholar] [CrossRef]
- Chen, J.; Chen, T.; Zhou, J.; Zhao, X.; Sheng, Q.; Lv, Z. MiR-146a-5p Mimic Inhibits NLRP3 Inflammasome Downstream Inflammatory Factors and CLIC4 in Neonatal Necrotizing Enterocolitis. Front. Cell Dev. Biol. 2021, 8, 594143. [Google Scholar] [CrossRef]
- Tong, J.; Duan, Z.; Zeng, R.; Du, L.; Xu, S.; Wang, L.; Liu, Y.; Chen, Q.; Chen, X.; Li, M. MiR-146a Negatively Regulates Aspergillus Fumigatus-Induced TNF-α and IL-6 Secretion in THP-1 Macrophages. Mycopathologia 2021, 186, 341–354. [Google Scholar] [CrossRef]
- Chen, L.; Ming, X.; Li, W.; Bi, M.; Yan, B.; Wang, X.; Yang, P.; Yang, B. The MicroRNA-155 Mediates Hepatitis B Virus Replication by Reinforcing SOCS1 Signalling–Induced Autophagy. Cell Biochem. Funct. 2020, 38, 436–442. [Google Scholar] [CrossRef]
- Babuta, M.; Furi, I.; Bala, S.; Bukong, T.N.; Lowe, P.; Catalano, D.; Calenda, C.; Kodys, K.; Szabo, G. Dysregulated Autophagy and Lysosome Function Are Linked to Exosome Production by Micro-RNA 155 in Alcoholic Liver Disease. Hepatology 2019, 70, 2123–2141. [Google Scholar] [CrossRef]
- Tsujimoto, T.; Mori, T.; Houri, K.; Onodera, Y.; Takehara, T.; Shigi, K.; Nakao, S.; Teramura, T.; Fukuda, K. MiR-155 Inhibits Mitophagy through Suppression of BAG5, a Partner Protein of PINK1. Biochem. Biophys. Res. Commun. 2020, 523, 707–712. [Google Scholar] [CrossRef]
- Criado, O.; Aguado, C.; Gayarre, J.; Duran-Trio, L.; Garcia-Cabrero, A.M.; Vernia, S.; San Millan, B.; Heredia, M.; Romá-Mateo, C.; Mouron, S.; et al. Lafora Bodies and Neurological Defects in Malin-Deficient Mice Correlate with Impaired Autophagy. Hum. Mol. Genet. 2012, 21, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Knecht, E.; Aguado, C.; Sarkar, S.; Korolchuk, V.I.; Criado-Garcia, O.; Vernia, S.; Boya, P.; Sanz, P.; Rodriguez de Cordoba, S.; Rubinsztein, D.C. Impaired Autophagy in Lafora Disease. Autophagy 2010, 6, 991–993. [Google Scholar] [CrossRef] [Green Version]
- Aguado, C.; Bovolenta, P.; Criado-García, O.; de Córdoba, S.R.; Dominguez, M.; Duran-Trio, L.; Garcia-Cabrero, A.M.; Gayarre, J.; Heredia, M.; Juana-López, L.; et al. Malin Knockout Mice Support a Primary Role of Autophagy in the Pathogenesis of Lafora Disease. Autophagy 2012, 8, 701–703. [Google Scholar] [CrossRef] [Green Version]
- Lahuerta, M.; Aguado, C.; Sánchez-Martín, P.; Sanz, P.; Knecht, E. Degradation of Altered Mitochondria by Autophagy Is Impaired in Lafora Disease. FEBS J. 2018, 285, 2071–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, P.K.; Roncon, P.; Lukasiuk, K.; Gorter, J.A.; Aronica, E.; Pitkänen, A.; Petretto, E.; Johnson, M.R.; Simonato, M. Meta-Analysis of MicroRNAs Dysregulated in the Hippocampal Dentate Gyrus of Animal Models of Epilepsy. eNeuro 2017, 4, ENEURO.0152-17.2017. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, D.; Peariso, K.; Gross, C. MicroRNA-Induced Silencing in Epilepsy: Opportunities and Challenges for Clinical Application. Dev. Dyn. 2018, 247, 94–110. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Chen, J.; Tao, H.; Cai, Y.; Huang, L.; Zhou, H.; Chen, Y.; Cui, L.; Zhong, W.; Li, K. Intranasal Delivery of MiR-155-5p Antagomir Alleviates Acute Seizures Likely by Inhibiting Hippocampal Inflammation. Neuropsychiatr Dis. Treat 2020, 16, 1295–1307. [Google Scholar] [CrossRef]
| Epm2a−/−vs. Control | Log2FC/FC | p-value | FDR |
| mmu-miR-155-5p | 3.81 | 1.366 × 10−7 | 8.933 × 10−5 |
| mmu-miR-146a-5p | 2.17 | 8.018 × 10−7 | 2.622 × 10−4 |
| Epm2b−/−vs. control | Log2FC/FC | p-value | FDR |
| mmu-miR-155-5p | 3.41 | 5.194 × 10−7 | 3.397 × 10−4 |
| mmu-miR-146a-5p | 2.13 | 1.138 × 10−6 | 3.720 × 10−4 |
| mmu-miR-10b-5p | 0.42 | 1.933 × 10−4 | 4.213 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romá-Mateo, C.; Lorente-Pozo, S.; Márquez-Thibaut, L.; Moreno-Estellés, M.; Garcés, C.; González, D.; Lahuerta, M.; Aguado, C.; García-Giménez, J.L.; Sanz, P.; et al. Age-Related microRNA Overexpression in Lafora Disease Male Mice Provides Links between Neuroinflammation and Oxidative Stress. Int. J. Mol. Sci. 2023, 24, 1089. https://doi.org/10.3390/ijms24021089
Romá-Mateo C, Lorente-Pozo S, Márquez-Thibaut L, Moreno-Estellés M, Garcés C, González D, Lahuerta M, Aguado C, García-Giménez JL, Sanz P, et al. Age-Related microRNA Overexpression in Lafora Disease Male Mice Provides Links between Neuroinflammation and Oxidative Stress. International Journal of Molecular Sciences. 2023; 24(2):1089. https://doi.org/10.3390/ijms24021089
Chicago/Turabian StyleRomá-Mateo, Carlos, Sheila Lorente-Pozo, Lucía Márquez-Thibaut, Mireia Moreno-Estellés, Concepción Garcés, Daymé González, Marcos Lahuerta, Carmen Aguado, José Luis García-Giménez, Pascual Sanz, and et al. 2023. "Age-Related microRNA Overexpression in Lafora Disease Male Mice Provides Links between Neuroinflammation and Oxidative Stress" International Journal of Molecular Sciences 24, no. 2: 1089. https://doi.org/10.3390/ijms24021089






