Specific Amino Acid Substitutions in OXA-51-Type β-Lactamase Enhance Catalytic Activity to a Level Comparable to Carbapenemase OXA-23 and OXA-24/40
Abstract
:1. Importance
2. Introduction
3. Materials and Methods
3.1. Antibiotics and Media
3.2. Genetic Analysis of OXA-51 Variants
3.3. Cloning of blaOXA and Site Directed Mutagenesis of blaOXA-51-Like Variants
3.4. OXA Type β-Lactamase Purification and Enzyme Kinetic Assay
3.5. Carbapenem Susceptibility Test of Constructs Carrying blaOXA Variants and Their Corresponding Mutants
3.6. Structure Modelling and Analysis
4. Results and Discussion
4.1. Genetic Analysis of Different OXA Variants
4.2. Enzymatic Activity of Different OXA Variants
4.3. Enzymatic Activity of Different OXA Mutants
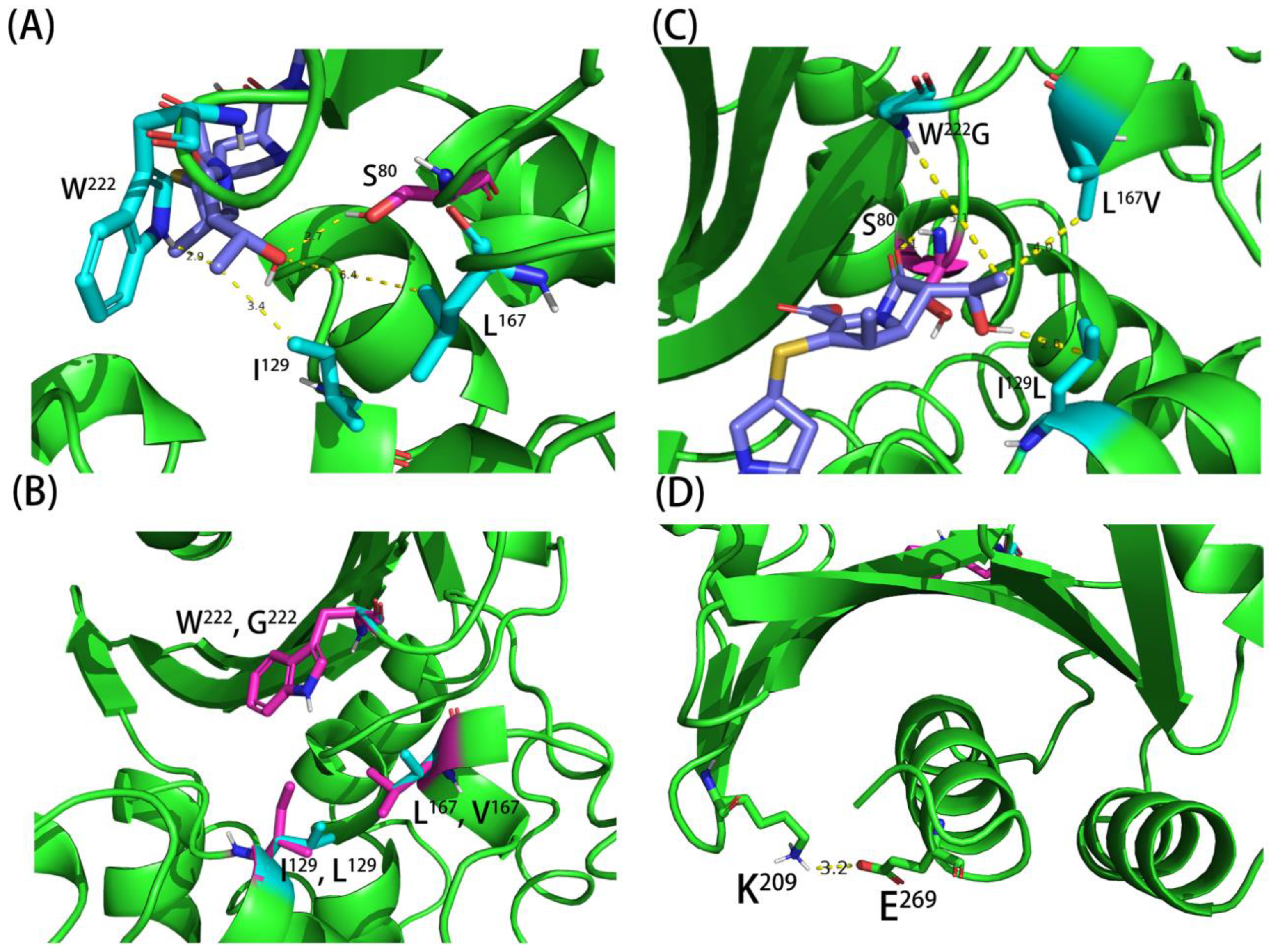
4.4. In Silico Modelling of Different OXA Mutants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bergogne-Bérézin, E.; Towner, K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Antibiotic Resistance Threats in the United States 2019; United States Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Charfi-Kessis, K.; Mansour, W.; Ben Haj Khalifa, A.; Mastouri, M.; Nordmann, P.; Aouni, M.; Poirel, L. Multidrug-resistant Acinetobacter baumannii strains carrying the bla(OxA-23) and the bla(GES-11) genes in a neonatology center in Tunisia. Microb. Pathog. 2014, 74, 20–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, W.W.; Kung, C.H.; Lee, C.H.; Tseng, C.P.; Wu, P.F.; Kuo, S.C.; Chen, T.L.; Lee, Y.T.; Wang, F.D.; Fung, C.P. Evolution of carbapenem resistance in Acinetobacter baumannii: An 18-year longitudinal study from a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 2015, 48, 57–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef] [Green Version]
- Savari, M.; Ekrami, A.; Shoja, S.; Bahador, A. Plasmid borne Carbapenem-Hydrolyzing Class D beta-Lactamases (CHDLs) and AdeABC efflux pump conferring carbapenem-tigecycline resistance among Acinetobacter baumannii isolates harboring TnAbaRs. Microb. Pathog. 2017, 104, 310–317. [Google Scholar] [CrossRef]
- Sari, A.N.; Bicmen, M.; Gulay, Z. The first report on the outbreak of OXA-24/40-like carbapenemase-producing Acinetobacter baumannii in Turkey. Jpn. J. Infect. Dis. 2013, 66, 439–442. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [Green Version]
- Higgins, P.G.; Pérez-Llarena, F.J.; Zander, E.; Fernández, A.; Bou, G.; Seifert, H. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 2121–2126. [Google Scholar] [CrossRef] [Green Version]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef] [Green Version]
- June, C.M.; Muckenthaler, T.J.; Schroder, E.C.; Klamer, Z.L.; Wawrzak, Z.; Powers, R.A.; Szarecka, A.; Leonard, D.A. The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity. Protein Sci. 2016, 25, 2152–2163. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.M.; Leonard, D.A. Common clinical substitutions enhance the carbapenemase activity of OXA-51-like class D beta-lactamases from Acinetobacter spp. Antimicrob. Agents Chemother. 2014, 58, 7015–7016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroder, E.C.; Klamer, Z.L.; Saral, A.; Sugg, K.A.; June, C.M.; Wymore, T.; Szarecka, A.; Leonard, D.A. Clinical Variants of the Native Class D beta-Lactamase of Acinetobacter baumannii Pose an Emerging Threat through Increased Hydrolytic Activity against Carbapenems. Antimicrob. Agents Chemother. 2016, 60, 6155–6164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zander, E.; Nemec, A.; Seifert, H.; Higgins, P.G. Association between β-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J. Clin. Microbiol. 2012, 50, 1900–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, B.A.; Brown, S.; Hamouda, A.; Findlay, J.; Amyes, S.G.B. Eleven novel OXA-51-like enzymes from clinical isolates of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007, 13, 1137–1138. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Namdari, F.; Réglier-Poupet, H.; Poyart, C.; Nordmann, P. Panresistant extended-spectrum β-lactamase SHV-5-producing Acinetobacter baumannii from New York City. J. Antimicrob. Chemother. 2007, 60, 1174–1176. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.H.-y.; Chan, B.K.-w.; Chan, E.W.-c.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ß-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [Green Version]
- Chiou, J.; Li, R.; Chen, S. CARB-17 family of beta-lactamases mediated intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 2015, 59, 3593–3595. [Google Scholar] [CrossRef] [Green Version]
- Chiou, J.; Leung, T.Y.; Chen, S. Molecular mechanisms of substrate recognition and specificity of New Delhi metallo-beta-lactamase. Antimicrob. Agents Chemother. 2014, 58, 5372–5378. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, J.H. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2015. [Google Scholar]
- Evans, B.A.; Hamouda, A.; Towner, K.J.; Amyes, S.G.B. OXA-51-like β-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin. Microbiol. Infect. 2008, 14, 268–275. [Google Scholar] [CrossRef]
- Takebayashi, Y.; Findlay, J.; Heesom, K.J.; Warburton, P.J.; Avison, M.B.; Evans, B.A. Variability in carbapenemase activity of intrinsic OxaAb (OXA-51-like) β-lactamase enzymes in Acinetobacter baumannii. J. Antimicrob. Chemother. 2020, 76, 587–595. [Google Scholar] [CrossRef]
- Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Frase, H.; Toth, M.; Kantardjieff, K.A.; Vakulenko, S. Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D β-Lactamases. ACS Chem. Biol. 2015, 10, 1791–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Primer | Sequence 5′ to 3 | Applications |
|---|---|---|
| OXA23-BamHI | CGATGGATCCATGAGTTATCTATTTTTGTCGTGTACAGAG | Cloning of blaOXA-23 |
| OXA23-NdeI | CGATCATATGTTAAATAATATTCAGCTGTTTTAATGATTTCATCAA | |
| OXA51-BamHI | CGATCGATCGGATCCAATCCAAATCACAGCGCTTCA | Cloning of blaOXA-51 and its variants |
| OXA51-NdeI | CGATCATATGCTATAAAATACCTAATTGTTCTAAGCTTTTATAAGT | |
| OXA72-BamHI | CGATGGATCCTCTATTAAAACTAAATCTGAAGATAATTTTCATATT | Cloning of blaOXA-72 |
| OXA72-NdeI | CGATCATATGTTAAATGATTCCAAGATTTTCTAGCG | |
| ISAba1-BamHI | CGATGGATCCCTAAATGATTGGTGACAATGAAGTTTTTTT | Cloning of blaOXA-23, blaOXA-51, and their variants (for Carbapenem susceptibility test) |
| blaOXA51-XhoI | CGATCTCGAGCTATAAAATACCTAATTGTTCTAAGCTTTTA | |
| blaOXA23-XhoI | CGATCTCGAGTTAAATAATATTCAGCTGTTTTAATGATTTCATCA | |
| blaOXA72-BamHI | CGATGGATCCCGATTCTTAGCCTCATCCCA | Cloning of blaOXA-72 (for Carbapenem susceptibility test) |
| blaOXA72-XhoI | CGATCTCGAGTTAAATGATTCCAAGATTTTCTAGCGACT | |
| Q57R-F | GTGTTTTAGTTATCCGACAAGGCCAAACTCA | Mutagenesis of Q57R |
| Q57R-R | TGAGTTTGGCCTTGTCGGATAACTAAAACAC | |
| I129L-F | ATGAAAGCTTCCGCTCTTCCAGTTTATCAAG | Mutagenesis of I129L |
| I129L-R | CTTGATAAACTGGAAGAGCGGAAGCTTTCAT | |
| L167V-F | GTCGATAATTTTTGGGTGGTGGGTCCTTTAA | Mutagenesis of L167V |
| L167V-R | TTAAAGGACCCACCACCCAAAAATTATCGAC | |
| K209M-F | TATTCATAGAAGAAATGAATGGAAACAAAAT | Mutagenesis of K209M |
| K209M-R | ATTTTGTTTCCATTCATTTCTTCTATGAATA | |
| W222G-F | AAAAGTGGTTGGGGAGGGGATGTAAACCCAC | Mutagenesis of W222G |
| W222G-R | GTGGGTTTACATCCCCTCCCCAACCACTTTT |
| OXA-23/OXA-24/OXA-51 Variants | Kinetics Constants | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Imipenem | Cefotaxime | ||||||||||||
| MIC (mg/L) | kcat (s−1) | km (μM) | kcat/km (μM−1/s−1) | Fold Increase | MIC (mg/L) | kcat (s−1) | km (μM) | kcat/km (μM−1/s−1) | Fold Increase | MIC (mg/L) | kcat (s−1) | Km (μM) | kcat/km (μM−1/s−1) | |
| OXA-23 | >2048 | 6.00 ±0.34 | 53.1 | 1.129 × 10−1 | 80.91 | 32 | 0.192 ± 0.006 | 0.88 | 2.174 × 10−1 | 39.06 | 8 | * n.d | n.d | n.d |
| OXA-72 | 1024 | 5.63 ± 0.53 | 198.4 | 2.839 × 10−2 | 20.34 | 64 | 0.145 ± 0.005 | 0.37 | 3.965 × 10−1 | 71.24 | 8 | n.d | n.d | n.d |
| OXA-51 | 32 | 1.32 ± 0.25 | 945.6 | 1.396 × 10−3 | 1.00 | 1 | 0.268 ± 0.014 | 48.2 | 5.566 × 10−3 | 1.00 | 8 | n.d | n.d | n.d |
| OXA-66 | 32 | 1.02 ± 0.37 | 1576 | 5.683 × 10−4 | 0.41 | 1 | 0.113 ± 0.004 | 34.3 | 3.291 × 10−3 | 0.59 | 8 | n.d | n.d | n.d |
| OXA-79 | >2048 | 16.61 ± 2.14 | 190.2 | 8.733 × 10−2 | 62.56 | 4 | 0.158 ± 0.003 | 2.9 | 5.533 × 10−2 | 9.94 | 8 | n.d | n.d | n.d |
| OXA-82 | 32 | 3.01 ± 0.53 | 619.9 | 4.854 × 10−3 | 3.48 | 16 | 0.226 ± 0.006 | 1.8 | 1.221 × 10−1 | 21.94 | 8 | n.d | n.d | n.d |
| OXA-83 | 32 | 0.87 ± 0.05 | 223.9 | 3.872 × 10−3 | 2.77 | 16 | 0.095 ± 0.001 | 0.63 | 1.517 × 10−1 | 27.25 | 8 | n.d | n.d | n.d |
| OXA-99 | 32 | 5.72 ± 2.27 | 2370 | 2.413 × 10−3 | 1.73 | 1 | 0.337 ± 0.020 | 33.8 | 9.973 × 10−3 | 1.79 | 8 | n.d | n.d | n.d |
| ATCC17978 | 8 | - | - | - | - | 0.125 | - | - | - | - | 8 | - | - | - |
| ATCC25922 | 4 | - | - | - | - | ≤0.06 | - | - | - | - | ≤0.06 | - | - | - |
| OXA-23/OXA-24/OXA-51 Variants | Amino Acid Substitutions in OXA-51/OXA-66 | Kinetic Constants | MIC (mg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | km (µM) | kcat/km (µM−1 s−1) | Fold Increase a | Biapenem | Meropenem | Imipenem | Ertapenem | ||
| OXA-23 | - | 0.015 ± 0.001 | 0.35 | 4.28 × 10−2 | 5.25 | 16 | 128 | 32 | 256 |
| OXA-72 b | - | 0.013 ± 0.0005 | 0.22 | 5.80 × 10−2 | 7.11 | 32 | 128 | 64 | 512 |
| OXA-66 | OXA-51 (T5A, E36V, V48A, Q107K, P194Q, D225N) | 0.03 ± 0.002 | 3.75 | 8.15 × 10−3 | 1.00 | 0.25 | 0.5 | 0.5 | 4 |
| OXA-66 (K209M) | OXA-66 (K209M) | 0.004 ± 0.0010 | 1.23 | 3.25 × 10−3 | 0.40 | 0.5 | 1 | 1 | 16 |
| OXA-79 | OXA-66 (W222G) | 0.018 ± 0.0008 | 0.72 | 2.50 × 10−2 | 3.07 | 4 | 16 | 4 | 128 |
| OXA-79 (I129L) | OXA-66 (W222G, I129L) | 0.007 ± 0.0010 | 0.15 | 4.67 × 10−2 | 5.72 | 8 | 32 | 8 | 256 |
| OXA-79 (K209M) | OXA-66 (W222G, K209M) | 0.016 ± 0.0008 | 1.21 | 1.3 × 10−2 | 1.37 | 2 | 8 | 2 | 32 |
| OXA-79 (W222M) | OXA-66 (W222M) | 0.010 ± 0.0009 | 0.18 | 5.55 × 10−2 | 5.86 | 16 | 64 | 8 | 256 |
| OXA-82 | OXA-66 (L167V) | 0.009 ± 0.0008 | 0.85 | 1.08 × 10−2 | 1.33 | 1 | 1 | 1 | 4 |
| OXA-82 (I129L) | OXA-66 (L167V, I129L) | 0.009 ± 0.0009 | 0.21 | 4.30 × 10−2 | 5.27 | 16 | 64 | 8 | 128 |
| OXA-82 (I129L) | OXA-66 (L167V, I129L) | - | - | - | - | 4 | 8 | 4 | 64 |
| OXA-82 (K209M) | OXA-66 (L167V, K209M) | 0.009 ± 0.0008 | 1.54 | 5.84 × 10−3 | 0.72 | 0.125 | 0.25 | 0.25 | 1 |
| OXA-82 (W222G) | OXA-66 (L167V, W222G) | 0.010 ± 0.0006 | 0.67 | 1.49 × 10−2 | 1.83 | 4 | 8 | 4 | 32 |
| OXA-82 (W222M) | OXA-66 (L167V, W222M) | 0.021 ± 0.0005 | 0.62 | 3.38 × 10−2 | 3.57 | 8 | 64 | 8 | 128 |
| OXA-82 (I129L, W222G) | OXA-66 (L167V, I129L, W222G) | 0.020 ± 0.0010 | 0.37 | 5.40 × 10−2 | 6.62 | 32 | 128 | 32 | 256 |
| OXA-83 | OXA-66 (I129L) | 0.018 ± 0.0004 | 0.49 | 3.67 × 10−2 | 4.50 | 8 | 128 | 32 | 256 |
| OXA-66 (I129A) | OXA-66 (I129A) | - | - | - | - | 8 | 32 | 8 | 256 |
| OXA-66 (I129D) | OXA-66 (I129D) | - | - | - | - | 1 | 2 | 1 | 32 |
| OXA-66 (I129F) | OXA-66 (I129F) | - | - | - | - | 4 | 16 | 4 | 128 |
| OXA-66 (I129M) | OXA-66 (I129M) | - | - | - | - | 2 | 16 | 4 | 64 |
| OXA-66 (I129V) | OXA-66 (I129V) | - | - | - | - | 8 | 32 | 8 | 256 |
| OXA-83 (W222G) | OXA-66 (I129L, W222G) | 0.023 ± 0.0011 | 0.39 | 5.89 × 10−2 | 7.22 | 8 | 16 | 8 | 256 |
| OXA-83 (W222M) | OXA-66 (I129L, W222M) | 0.042 ± 0.0011 | 0.36 | 1.16 × 10−1 | 12.27 | 64 | 256 | 128 | >512 |
| OXA-99 | OXA-51 (Q57R, K209M) | 0.019 ± 0.0009 | 2.7 | 7.03 × 10−3 | 0.86 | 0.125 | 0.25 | 0.25 | 4 |
| OXA-99 (I129L) | OXA-51 (Q57R, K209M, I129L) | 0.021 ± 0.0007 | 1.63 | 1.29 × 10−2 | 1.58 | 2 | 4 | 2 | 32 |
| Control c | - | - | - | - | - | ≤0.06 | 0.125 | 0.125 | 0.125 |
| Control d | - | - | - | - | - | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, K.-W.; Liu, C.-Y.; Wong, H.-Y.; Chan, W.-C.; Wong, K.-Y.; Chen, S. Specific Amino Acid Substitutions in OXA-51-Type β-Lactamase Enhance Catalytic Activity to a Level Comparable to Carbapenemase OXA-23 and OXA-24/40. Int. J. Mol. Sci. 2022, 23, 4496. https://doi.org/10.3390/ijms23094496
Chan K-W, Liu C-Y, Wong H-Y, Chan W-C, Wong K-Y, Chen S. Specific Amino Acid Substitutions in OXA-51-Type β-Lactamase Enhance Catalytic Activity to a Level Comparable to Carbapenemase OXA-23 and OXA-24/40. International Journal of Molecular Sciences. 2022; 23(9):4496. https://doi.org/10.3390/ijms23094496
Chicago/Turabian StyleChan, Kwan-Wai, Chen-Yu Liu, Ho-Yin Wong, Wai-Chi Chan, Kwok-Yin Wong, and Sheng Chen. 2022. "Specific Amino Acid Substitutions in OXA-51-Type β-Lactamase Enhance Catalytic Activity to a Level Comparable to Carbapenemase OXA-23 and OXA-24/40" International Journal of Molecular Sciences 23, no. 9: 4496. https://doi.org/10.3390/ijms23094496






