Roles of Mitochondrial DNA Damage in Kidney Diseases: A New Biomarker
Abstract
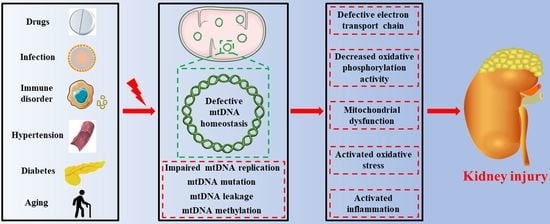
:1. Introduction
2. Common Types of mtDNA Damage
2.1. Impaired mtDNA Replication
2.2. mtDNA Mutations
2.3. mtDNA Leakage
2.4. mtDNA Methylation
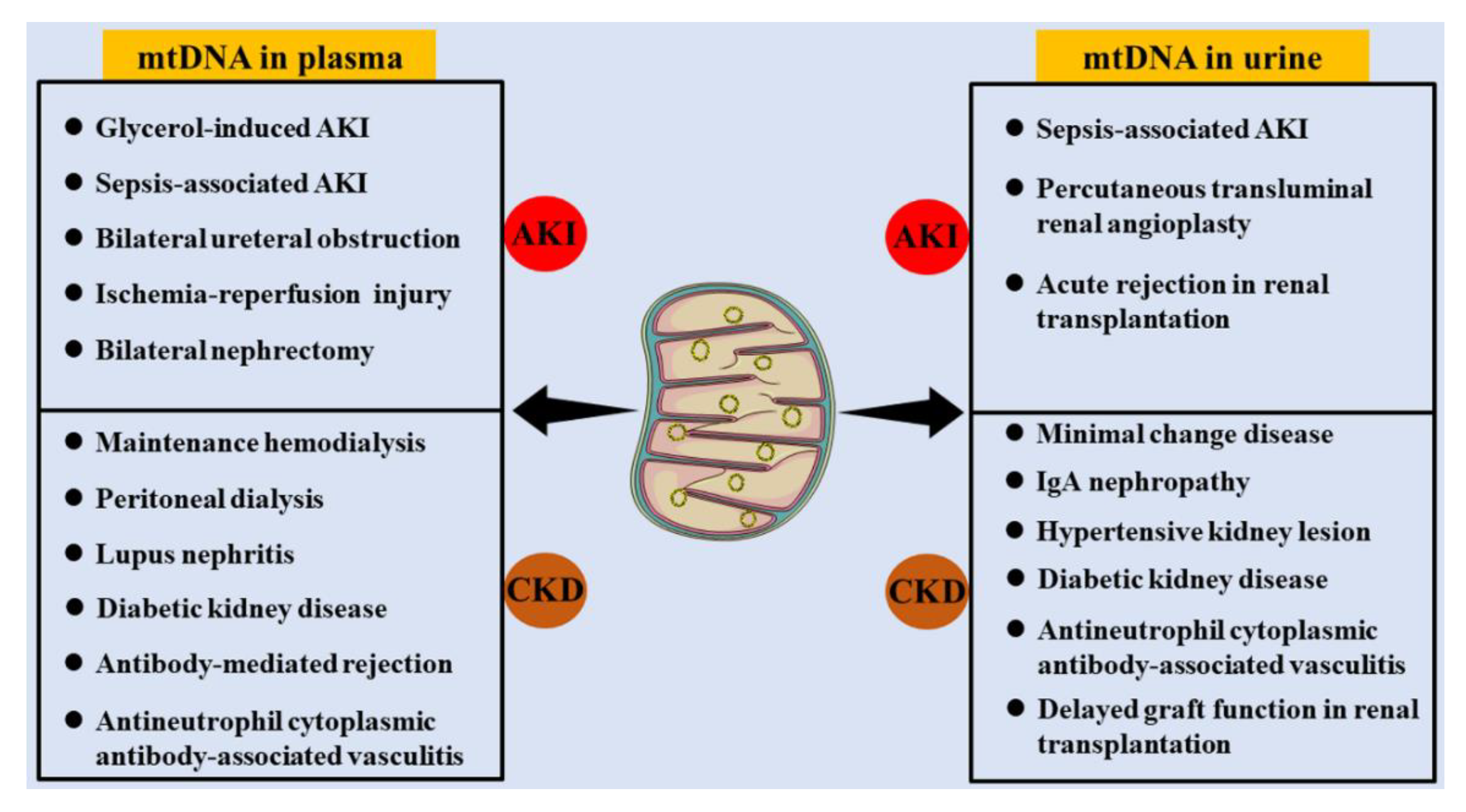
3. mtDNA Distribution in Kidney Diseases
3.1. mtDNA in Peripheral Serum
3.2. mtDNA in Urine
4. mtDNA Damage in Kidney Diseases
4.1. Impaired mtDNA Replication
4.2. mtDNA Mutations
4.3. mtDNA Leakage
4.4. mtDNA Methylation
5. Pharmacological Intervention of mtDNA Damage in Kidney Diseases
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, C.; Cai, J.; Yin, X.M.; Weinberg, J.M.; Venkatachalam, M.A.; Dong, Z. Mitochondrial quality control in kidney injury and repair. Nat. Rev. Nephrol. 2021, 17, 299–318. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Capili, A.; Choi, M.E. Mitochondrial dysfunction in kidney injury, inflammation, and disease: Potential therapeutic approaches. Kidney Res. Clin. Pr. 2020, 39, 244–258. [Google Scholar] [CrossRef]
- Gilea, A.I.; Ceccatelli, B.C.; Magistrati, M.; di Punzio, G.; Goffrini, P.; Baruffini, E.; Dallabona, C. Saccharomyces cerevisiae as a Tool for Studying Mutations in Nuclear Genes Involved in Diseases Caused by Mitochondrial DNA Instability. Genes 2021, 12, 1866. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, K.A.; Rooney, J.P.; Turner, E.A.; Donoghue, L.J.; Bodhicharla, R.; Maurer, L.L.; Ryde, I.T.; Kim, J.J.; Joglekar, R.; Hibshman, J.D.; et al. Early-life mitochondrial DNA damage results in lifelong deficits in energy production mediated by redox signaling in Caenorhabditis elegans. Redox Biol. 2021, 43, 102000. [Google Scholar] [CrossRef] [PubMed]
- van der Slikke, E.C.; Star, B.S.; van Meurs, M.; Henning, R.H.; Moser, J.; Bouma, H.R. Sepsis is associated with mitochondrial DNA damage and a reduced mitochondrial mass in the kidney of patients with sepsis-AKI. Crit. Care 2021, 25, 36. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Yu, B.; Armando, I.; Han, F. Mitochondrial DNA-Mediated Inflammation in Acute Kidney Injury and Chronic Kidney Disease. Oxid Med. Cell Longev. 2021, 2021, 9985603. [Google Scholar] [CrossRef]
- Melki, I.; Allaeys, I.; Tessandier, N.; Levesque, T.; Cloutier, N.; Laroche, A.; Vernoux, N.; Becker, Y.; Benk-Fortin, H.; Zufferey, A.; et al. Platelets release mitochondrial antigens in systemic lupus erythematosus. Sci. Transl. Med. 2021, 13, eaav5928. [Google Scholar] [CrossRef]
- Kockler, Z.W.; Osia, B.; Lee, R.; Musmaker, K.; Malkova, A. Repair of DNA Breaks by Break-Induced Replication. Annu. Rev. Biochem. 2021, 90, 165–191. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, X.; Hu, Q.; Wu, J.; Wang, G.; Hong, Z.; Ren, J. Mitochondrial DNA in liver inflammation and oxidative stress. Life Sci. 2019, 236, 116464. [Google Scholar] [CrossRef]
- Lin, Y.H.; Lim, S.N.; Chen, C.Y.; Chi, H.C.; Yeh, C.T.; Lin, W.R. Functional Role of Mitochondrial DNA in Cancer Progression. Int. J. Mol. Sci. 2022, 23, 1659. [Google Scholar] [CrossRef]
- Lechuga-Vieco, A.V.; Latorre-Pellicer, A.; Calvo, E.; Torroja, C.; Pellico, J.; Acin-Perez, R.; Garcia-Gil, M.L.; Santos, A.; Bagwan, N.; Bonzon-Kulichenko, E.; et al. Heteroplasmy of Wild-Type Mitochondrial DNA Variants in Mice Causes Metabolic Heart Disease With Pulmonary Hypertension and Frailty. Circulation 2022, 145, 1084–1101. [Google Scholar] [CrossRef]
- Zhong, W.; Rao, Z.; Xu, J.; Sun, Y.; Hu, H.; Wang, P.; Xia, Y.; Pan, X.; Tang, W.; Chen, Z.; et al. Defective mitophagy in aged macrophages promotes mitochondrial DNA cytosolic leakage to activate STING signaling during liver sterile inflammation. Aging Cell 2022, 21, e13622. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Murley, A.; Golder, Z.; Rowe, J.B.; Allinson, K.; Chinnery, P.F. Heteroplasmic mitochondrial DNA mutations in frontotemporal lobar degeneration. Acta Neuropathol. 2022, 143, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Stoccoro, A.; Coppede, F. Mitochondrial DNA Methylation and Human Diseases. Int. J. Mol. Sci. 2021, 22, 4594. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, T.; Setoyama, D.; Yasukawa, T.; Kang, D. Mitochondria metabolomics reveals a role of beta-nicotinamide mononucleotide metabolism in mitochondrial DNA replication. J. Biochem. 2022, 171, 325–338. [Google Scholar] [CrossRef]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef]
- Roy, A.; Kandettu, A.; Ray, S.; Chakrabarty, S. Mitochondrial DNA replication and repair defects: Clinical phenotypes and therapeutic interventions. Biochim. Biophys. Acta Bioenerg. 2022, 1863, 148554. [Google Scholar] [CrossRef]
- Manini, A.; Abati, E.; Comi, G.P.; Corti, S.; Ronchi, D. Mitochondrial DNA homeostasis impairment and dopaminergic dysfunction: A trembling balance. Ageing Res. Rev. 2022, 76, 101578. [Google Scholar] [CrossRef]
- Blazquez-Bermejo, C.; Carreno-Gago, L.; Molina-Granada, D.; Aguirre, J.; Ramon, J.; Torres-Torronteras, J.; Cabrera-Perez, R.; Martin, M.A.; Dominguez-Gonzalez, C.; de la Cruz, X.; et al. Increased dNTP pools rescue mtDNA depletion in human POLG-deficient fibroblasts. FASEB J. 2019, 33, 7168–7179. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, P.; Anderson, N.S.; Shpilka, T.; Du, Y.; Naresh, N.U.; Li, R.; Zhu, L.J.; Luk, K.; Lavelle, J.; et al. LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria. Nat. Cell Biol. 2022, 24, 181–193. [Google Scholar] [CrossRef]
- Piro-Megy, C.; Sarzi, E.; Tarres-Sole, A.; Pequignot, M.; Hensen, F.; Quiles, M.; Manes, G.; Chakraborty, A.; Senechal, A.; Bocquet, B.; et al. Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy. J. Clin. Investig. 2020, 130, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Xie, X.; Zhu, X.; Jiang, S.; Milenkovic, D.; Misic, J.; Shi, Y.; Tandukar, N.; Li, X.; Atanassov, I.; et al. The mitochondrial single-stranded DNA binding protein is essential for initiation of mtDNA replication. Sci. Adv. 2021, 7, eabf8631. [Google Scholar] [CrossRef] [PubMed]
- Otten, A.; Kamps, R.; Lindsey, P.; Gerards, M.; Pendeville-Samain, H.; Muller, M.; van Tienen, F.; Smeets, H. Tfam Knockdown Results in Reduction of mtDNA Copy Number, OXPHOS Deficiency and Abnormalities in Zebrafish Embryos. Front. Cell Dev. Biol 2020, 8, 381. [Google Scholar] [CrossRef] [PubMed]
- Sercel, A.J.; Carlson, N.M.; Patananan, A.N.; Teitell, M.A. Mitochondrial DNA Dynamics in Reprogramming to Pluripotency. Trends Cell Biol. 2021, 31, 311–323. [Google Scholar] [CrossRef]
- Sato, T.; Goto-Inoue, N.; Kimishima, M.; Toyoharu, J.; Minei, R.; Ogura, A.; Nagoya, H.; Mori, T. A novel ND1 mitochondrial DNA mutation is maternally inherited in growth hormone transgenesis in amago salmon (Oncorhynchus masou ishikawae). Sci. Rep. 2022, 12, 6720. [Google Scholar] [CrossRef]
- McMillan, R.P.; Stewart, S.; Budnick, J.A.; Caswell, C.C.; Hulver, M.W.; Mukherjee, K.; Srivastava, S. Quantitative Variation in m.3243A > G Mutation Produce Discrete Changes in Energy Metabolism. Sci. Rep. 2019, 9, 5752. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Liu, X.; Zhou, J.; Li, T. Mitochondrial DNA Is a Vital Driving Force in Ischemia-Reperfusion Injury in Cardiovascular Diseases. Oxid. Med. Cell Longev. 2022, 2022, 6235747. [Google Scholar] [CrossRef]
- Xu, C.; Tong, L.; Rao, J.; Ye, Q.; Chen, Y.; Zhang, Y.; Xu, J.; Mao, X.; Meng, F.; Shen, H.; et al. Heteroplasmic and homoplasmic m.616T>C in mitochondria tRNAPhe promote isolated chronic kidney disease and hyperuricemia. JCI Insight. 2022, 7, e157418. [Google Scholar] [CrossRef]
- Ji, X.; Guo, W.; Gu, X.; Guo, S.; Zhou, K.; Su, L.; Yuan, Q.; Liu, Y.; Guo, X.; Huang, Q.; et al. Mutational profiling of mtDNA control region reveals tumor-specific evolutionary selection involved in mitochondrial dysfunction. Ebiomedicine 2022, 80, 104058. [Google Scholar] [CrossRef]
- Nunn, C.J.; Goyal, S. Contingency and selection in mitochondrial genome dynamics. Elife 2022, 11, e76557. [Google Scholar] [CrossRef]
- Hancock-Cerutti, W.; Wu, Z.; Xu, P.; Yadavalli, N.; Leonzino, M.; Tharkeshwar, A.K.; Ferguson, S.M.; Shadel, G.S.; De Camilli, P. ER-lysosome lipid transfer protein VPS13C/PARK23 prevents aberrant mtDNA-dependent STING signaling. J. Cell Biol. 2022, 221, e202106046. [Google Scholar] [CrossRef] [PubMed]
- McArthur, K.; Whitehead, L.W.; Heddleston, J.M.; Li, L.; Padman, B.S.; Oorschot, V.; Geoghegan, N.D.; Chappaz, S.; Davidson, S.; San, C.H.; et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 2018, 359, eaao6047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Gupta, R.; Blanco, L.P.; Yang, S.; Shteinfer-Kuzmine, A.; Wang, K.; Zhu, J.; Yoon, H.E.; Wang, X.; Kerkhofs, M.; et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 2019, 366, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Davidson, S.; Harapas, C.R.; Hilton, J.B.; Mlodzianoski, M.J.; Laohamonthonkul, P.; Louis, C.; Low, R.; Moecking, J.; De Nardo, D.; et al. TDP-43 Triggers Mitochondrial DNA Release via mPTP to Activate cGAS/STING in ALS. Cell 2020, 183, 636–649. [Google Scholar] [CrossRef]
- Li, J.S.; Hao, Y.Z.; Hou, M.L.; Zhang, X.; Zhang, X.G.; Cao, Y.X.; Li, J.M.; Ma, J.; Zhou, Z.X. Development of a Recombinase-aided Amplification Combined With Lateral Flow Dipstick Assay for the Rapid Detection of the African Swine Fever Virus. Biomed. Environ. Sci. 2022, 35, 133–140. [Google Scholar]
- Harapas, C.R.; Idiiatullina, E.; Al-Azab, M.; Hrovat-Schaale, K.; Reygaerts, T.; Steiner, A.; Laohamonthonkul, P.; Davidson, S.; Yu, C.H.; Booty, L.; et al. Organellar homeostasis and innate immune sensing. Nat. Rev. Immunol. 2022, 9, 539–545. [Google Scholar] [CrossRef]
- Luecke, S.; Holleufer, A.; Christensen, M.H.; Jonsson, K.L.; Boni, G.A.; Sorensen, L.K.; Johannsen, M.; Jakobsen, M.R.; Hartmann, R.; Paludan, S.R. cGAS is activated by DNA in a length-dependent manner. Embo. Rep. 2017, 18, 1707–1715. [Google Scholar] [CrossRef]
- Kato, K.; Omura, H.; Ishitani, R.; Nureki, O. Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu. Rev. Biochem. 2017, 86, 541–566. [Google Scholar] [CrossRef]
- Luteijn, R.D.; Zaver, S.A.; Gowen, B.G.; Wyman, S.K.; Garelis, N.E.; Onia, L.; McWhirter, S.M.; Katibah, G.E.; Corn, J.E.; Woodward, J.J.; et al. SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 2019, 573, 434–438. [Google Scholar] [CrossRef]
- Ding, P.; Tan, Q.; Wei, Z.; Chen, Q.; Wang, C.; Qi, L.; Wen, L.; Zhang, C.; Yao, C. Toll-like receptor 9 deficiency induces osteoclastic bone loss via gut microbiota-associated systemic chronic inflammation. Bone Res. 2022, 10, 42. [Google Scholar] [CrossRef]
- Honke, N.; Lowin, T.; Opgenoorth, B.; Shaabani, N.; Lautwein, A.; Teijaro, J.R.; Schneider, M.; Pongratz, G. Endogenously produced catecholamines improve the regulatory function.n of TLR9-activated B cells. PLoS Biol. 2022, 20, e3001513. [Google Scholar] [CrossRef] [PubMed]
- Hepokoski, M.; Singh, P. Mitochondria as mediators of systemic inflammation and organ cross talk in acute kidney injury. Am. J. Physiol. Ren. Physiol. 2022, 6, F589–F596. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.; Tsuji, T.; Ohashi, N.; Kato, A.; Fujigaki, Y.; Yasuda, H. Role of Mitochondrial DNA in Septic AKI via Toll-Like Receptor. J. Am. Soc. Nephrol. 2016, 27, 2009–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, P.; Toy, R.; Jhita, N.; Atalis, A.; Pandey, B.; Beach, A.; Blanchard, E.L.; Moore, S.G.; Gaul, D.A.; Santangelo, P.J.; et al. TRAF6-IRF5 kinetics, TRIF, and biophysical factors drive synergistic innate responses to particle-mediated MPLA-CpG co-presentation. Sci. Adv. 2021, 7, eabd4235. [Google Scholar] [CrossRef] [PubMed]
- De Gaetano, A.; Solodka, K.; Zanini, G.; Selleri, V.; Mattioli, A.V.; Nasi, M.; Pinti, M. Molecular Mechanisms of mtDNA-Mediated Inflammation. Cells 2021, 10, 2898. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, J.H.; Zhang, H.; Canfran-Duque, A.; Singh, A.K.; Perry, R.J.; Shulman, G.I.; Fernandez-Hernando, C.; Min, W. Brown adipose TRX2 deficiency activates mtDNA-NLRP3 to impair thermogenesis and protect against diet-induced insulin resistance. J. Clin. Invest. 2022, 132, e148852. [Google Scholar] [CrossRef]
- Wang, L.; Liu, T.; Yang, S.; Sun, L.; Zhao, Z.; Li, L.; She, Y.; Zheng, Y.; Ye, X.; Bao, Q.; et al. Perfluoroalkyl substance pollutants activate the innate immune system through the AIM2 inflammasome. Nat. Commun. 2021, 12, 2915. [Google Scholar] [CrossRef]
- Shen, J.; Wang, C.; Li, D.; Xu, T.; Myers, J.; Ashton, J.M.; Wang, T.; Zuscik, M.J.; McAlinden, A.; O’Keefe, R.J. DNA methyltransferase 3b regulates articular cartilage homeostasis by altering metabolism. JCI Insight. 2017, 2, e93612. [Google Scholar] [CrossRef] [Green Version]
- Damal, V.S.; Ebert, S.M.; Lim, H.W.; Kim, J.; You, D.; Jung, B.C.; Palacios, H.H.; Tcheau, T.; Adams, C.M.; Kang, S. A necessary role of DNMT3A in endurance exercise by suppressing.g ALDH1L1-mediated oxidative stress. Embo. J. 2021, 40, e106491. [Google Scholar]
- Liu, Q.; Li, H.; Guo, L.; Chen, Q.; Gao, X.; Li, P.H.; Tang, N.; Guo, X.; Deng, F.; Wu, S. Effects of short-term personal exposure to air pollution on platelet mitochondrial DNA methylation levels and the potential mitigation by L-arginine supplementation. J. Hazard. Mater. 2021, 417, 125963. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, J.; Wu, F.; Lai, C.; Li, Y.; Zhang, G.; Peng, X.; Yu, S.; Yang, J.; Wang, W.; et al. Biological and epigenetic alterations of mitochondria involved in cellular replicative and hydrogen peroxide-induced premature senescence of human embryonic lung fibroblasts. Ecotoxicol. Env. Saf 2021, 216, 112204. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Lv, W.; Wang, X.; Dong, S.; Liu, X.; Yang, T.; Xu, J.; Zeng, M.; Zou, X.; Zhao, D.; et al. Hypermethylation of Hepatic Mitochondrial ND6 Provokes Systemic Insulin Resistance. Adv. Sci (Weinh) 2021, 8, 2004507. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, F.; Yang, Y.; Yang, S.; Jiang, M.; Zhang, W.; Ma, Z.; Gu, X. Mitochondrial DNA methylation drift and postoperative delirium in mice. Eur. J. Anaesthesiol. 2022, 39, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shang, J.; Wang, F.; Huo, X.; Sun, R.; Ren, Z.; Wang, W.; Yang, M.; Li, G.; Gao, D.; et al. Decreased mitochondrial D-loop region methylation mediates an increase in mitochondrial DNA copy number in CADASIL. Clin. Epigenetics 2022, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Cushen, S.C.; Ricci, C.A.; Bradshaw, J.L.; Silzer, T.; Blessing, A.; Sun, J.; Zhou, Z.; Scroggins, S.M.; Santillan, M.K.; Santillan, D.A.; et al. Reduced Maternal Circulating Cell-Free Mitochondrial DNA Is Associated With the Development of Preeclampsia. J. Am. Heart Assoc. 2022, 11, e21726. [Google Scholar] [CrossRef]
- Wei, R.; Ni, Y.; Bazeley, P.; Grandhi, S.; Wang, J.; Li, S.T.; Hazen, S.L.; Wilson, T.W.; LaFramboise, T. Mitochondrial DNA Content Is Linked to Cardiovascular Disease Patient Phenotypes. J. Am. Heart Assoc. 2021, 10, e18776. [Google Scholar] [CrossRef]
- Zhong, W.; Rao, Z.; Rao, J.; Han, G.; Wang, P.; Jiang, T.; Pan, X.; Zhou, S.; Zhou, H.; Wang, X. Aging aggravated liver ischemia and reperfusion injury by promoting STING-mediated NLRP3 activation in macrophages. Aging Cell 2020, 19, e13186. [Google Scholar] [CrossRef]
- Gonzalez-Freire, M.; Moore, A.Z.; Peterson, C.A.; Kosmac, K.; McDermott, M.M.; Sufit, R.L.; Guralnik, J.M.; Polonsky, T.; Tian, L.; Kibbe, M.R.; et al. Associations of Peripheral Artery Disease With Calf Skeletal Muscle Mitochondrial DNA Heteroplasmy. J. Am. Heart Assoc. 2020, 9, e15197. [Google Scholar] [CrossRef]
- Homolova, J.; Janovicova, L.; Konecna, B.; Vlkova, B.; Celec, P.; Tothova, L.; Babickova, J. Plasma Concentrations of Extracellular DNA in Acute Kidney Injury. Diagnostics 2020, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Ren, J.; Liu, Q.; Hu, Q.; Wu, X.; Wang, G.; Hong, Z.; Ren, H.; Li, J. Effects of Changes in the Levels of Damage-Associated Molecular Patterns Following Continuous Veno-Venous Hemofiltration Therapy on Outcomes in Acute Kidney Injury Patients With Sepsis. Front. Immunol. 2018, 9, 3052. [Google Scholar] [CrossRef] [Green Version]
- Jancuska, A.; Potocarova, A.; Kovalcikova, A.G.; Podracka, L.; Babickova, J.; Celec, P.; Tothova, L. Dynamics of Plasma and Urinary Extracellular DNA in Acute Kidney Injury. Int. J. Mol. Sci. 2022, 23, 3402. [Google Scholar] [CrossRef] [PubMed]
- He, W.J.; Li, C.; Huang, Z.; Geng, S.; Rao, V.S.; Kelly, T.N.; Hamm, L.L.; Grams, M.E.; Arking, D.E.; Appel, L.J.; et al. Association of Mitochondrial DNA Copy Number with Risk of Progression of Kidney Disease. Clin. J. Am. Soc. Nephrol. 2022, 17, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wu, J.; Luo, J.; Chen, X.; Yang, J.; Fang, L. Urinary mitochondrial DNA: A potential early biomarker of diabetic nephropathy. Diabetes Metab. Res. Rev. 2019, 35, e3131. [Google Scholar] [CrossRef] [PubMed]
- Szeto, C.C.; Lai, K.B.; Chow, K.M.; Kwan, B.C.; Cheng, P.M.; Kwong, V.W.; Choy, A.S.; Leung, C.B.; Li, P.K. Plasma Mitochondrial DNA Level is a Prognostic Marker in Peritoneal Dialysis Patients. Kidney Blood Press Res. 2016, 41, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Guo, Y.; Zhong, X.Y. Circulating Cell-Free Mi.itochondrial DNA: A Potential Blood-Based Biomarker for Sarcopenia in Patients Undergoing Maintenance Hemodialysis. Med. Sci. Monit. 2022, 28, e934679. [Google Scholar]
- Tian, S.L.; Bai, X.; Xu, P.C.; Chen, T.; Gao, S.; Hu, S.Y.; Wei, L.; Jia, J.Y.; Yan, T.K. Circulating nicotinamide adenine dinucleotide-ubiquinone oxidoreductase chain 6 is associated with disease activity of anti-neutrophil cytoplasmic antibody-associated vasculitis. Clin. Chim. Acta 2020, 511, 125–131. [Google Scholar] [CrossRef]
- Han, F.; Sun, Q.; Huang, Z.; Li, H.; Ma, M.; Liao, T.; Luo, Z.; Zheng, L.; Zhang, N.; Chen, N.; et al. Donor plasma mitochondrial DNA is associated with antibody-mediated rejection in renal allograft recipients. Aging (Albany NY) 2021, 13, 8440–8453. [Google Scholar] [CrossRef]
- Han, F.; Wan, S.; Sun, Q.; Chen, N.; Li, H.; Zheng, L.; Zhang, N.; Huang, Z.; Hong, L.; Sun, Q. Donor Plasma Mitochondrial DNA Is Correlated with Posttransplant Renal Allograft Function. Transplantation 2019, 103, 2347–2358. [Google Scholar] [CrossRef]
- Eirin, A.; Herrmann, S.M.; Saad, A.; Abumoawad, A.; Tang, H.; Lerman, A.; Textor, S.C.; Lerman, L.O. Urinary mitochondrial DNA copy number identifies renal mitochondrial injury in renovascular hypertensive patients undergoing renal revascularization: A Pilot Study. Acta Physiol. (Oxf) 2019, 226, e13267. [Google Scholar] [CrossRef]
- Hu, Q.; Ren, J.; Ren, H.; Wu, J.; Wu, X.; Liu, S.; Wang, G.; Gu, G.; Guo, K.; Li, J. Urinary Mitochondrial DNA Identifies Renal Dysfunction and Mitochondrial Damage in Sepsis-Induced Acute Kidney Injury. Oxid Med. Cell Longev. 2018, 2018, 8074936. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.C.; Moon, A.; Lee, K.H.; Oh, Y.S.; Park, M.Y.; Choi, S.J.; Kim, J.K. Minimal Change Disease Is Associated with Mitochondrial Injury and STING Pathway Activation. J. Clin. Med. 2022, 11, 577. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.Z.; Kwan, B.C.; Chow, K.M.; Cheng, P.M.; Luk, C.C.; Li, P.K.; Szeto, C.C. Urinary mitochondrial DNA level is an indicator of intra-renal mitochondrial depletion and renal scarring in diabetic nephropathy. Nephrol. Dial. Transpl. 2018, 33, 784–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, P.Z.; Kwan, B.C.; Chow, K.M.; Cheng, P.M.; Luk, C.C.; Lai, K.B.; Li, P.K.; Szeto, C.C. Urinary mitochondrial DNA level in non-diabetic chronic kidney diseases. Clin. Chim. Acta 2018, 484, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chiu, P.F.; Wu, C.L.; Kuo, C.L.; Huang, C.S.; Liu, C.S.; Huang, C.H. Urinary cell-free mitochondrial and nuclear deoxyribonucleic acid correlates with the prognosis of chronic kidney diseases. BMC Nephrol. 2019, 20, 391. [Google Scholar] [CrossRef]
- Eirin, A.; Saad, A.; Tang, H.; Herrmann, S.M.; Woollard, J.R.; Lerman, A.; Textor, S.C.; Lerman, L.O. Urinary Mitochondrial DNA Copy Number Identifies Chronic Renal Injury in Hypertensive Patients. Hypertension 2016, 68, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Eirin, A.; Saad, A.; Woollard, J.R.; Juncos, L.A.; Calhoun, D.A.; Tang, H.; Lerman, A.; Textor, S.C.; Lerman, L.O. Glomerular Hyperfiltration in Obese African American Hypertensive Patients Is Associated With Elevated Urinary Mitochondrial-DNA Copy Number. Am. J. Hypertens. 2017, 30, 1112–1119. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.C.; Cho, N.J.; Park, S.; Kim, H.; Gil, H.W.; Lee, E.Y.; Kwon, S.H.; Jeon, J.S.; Noh, H.; Han, D.C.; et al. Minor Glomerular Abnormalities are Associated with Deterioration of Long-Term Kidney Function and Mitochondrial Injury. J. Clin. Med. 2019, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.C.; Cho, N.J.; Park, S.; Kim, H.; Choi, S.J.; Kim, J.K.; Hwang, S.D.; Gil, H.W.; Lee, E.Y.; Jeon, J.S.; et al. IgA nephropathy is associated with elevated urinary mitochondrial DNA copy numbers. Sci. Rep. 2019, 9, 16068. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.J.; Yang, X.; Xu, P.C.; Chen, T.; Gao, S.; Hu, S.Y.; Wei, L.; Yan, T.K. Urinary mitochondrial DNA is a useful biomarker for assessing kidney injury of antineutrophil cytoplasmic antibody -associated vasculitis. Clin. Chim. Acta 2020, 502, 263–268. [Google Scholar] [CrossRef]
- Jansen, M.; Pulskens, W.; Uil, M.; Claessen, N.; Nieuwenhuizen, G.; Standaar, D.; Hau, C.M.; Nieuwland, R.; Florquin, S.; Bemelman, F.J.; et al. Urinary mitochondrial DNA associates with delayed graft function following renal transplantation. Nephrol. Dial. Transpl. 2020, 35, 1320–1327. [Google Scholar] [CrossRef]
- Kim, K.; Moon, H.; Lee, Y.H.; Seo, J.W.; Kim, Y.G.; Moon, J.Y.; Kim, J.S.; Jeong, K.H.; Lee, T.W.; Ihm, C.G.; et al. Clinical relevance of cell-free mitochondrial DNA during the early postoperative period in kidney transplant recipients. Sci. Rep. 2019, 9, 18607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbers, E.; Kekalainen, N.J.; Hangas, A.; Pohjoismaki, J.L.; Goffart, S. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion 2019, 44, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Longchamps, R.J.; Yang, S.Y.; Castellani, C.A.; Shi, W.; Lane, J.; Grove, M.L.; Bartz, T.M.; Sarnowski, C.; Liu, C.; Burrows, K.; et al. Genome-wide analysis of mitochondrial DNA copy number reveals loci implicated in nucleotide metabolism, platelet activation, and megakaryocyte proliferation. Hum. Genet. 2022, 141, 127–146. [Google Scholar] [CrossRef]
- Gustafson, M.A.; McCormick, E.M.; Perera, L.; Longley, M.J.; Bai, R.; Kong, J.; Dulik, M.; Shen, L.; Goldstein, A.C.; McCormack, S.E.; et al. Mitochondrial single-stranded DNA binding protein novel de novo SSBP1 mutation in a child with single large-scale mtDNA deletion (SLSMD) clinically manifesting as Pearson, Kearns-Sayre, and Leigh syndromes. PLoS ONE 2019, 14, e221829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del, D.V.; Ullah, F.; Di Meo, I.; Magini, P.; Gusic, M.; Maresca, A.; Caporali, L.; Palombo, F.; Tagliavini, F.; Baugh, E.H.; et al. SSBP1 mutations cause mtDNA depletion underlying a complex optic atrophy disorder. J. Clin. Investig. 2020, 130, 108–125. [Google Scholar]
- Gureev, A.P.; Andrianova, N.V.; Pevzner, I.B.; Zorova, L.D.; Chernyshova, E.V.; Sadovnikova, I.S.; Chistyakov, D.V.; Popkov, V.A.; Semenovich, D.S.; Babenko, V.A.; et al. Dietary restriction modulates mitochondrial DNA damage and oxylipin profile in aged rats. FEBS J. 2022, 289, 5697–5713. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, Q.; Peiffer, L.B.; Hicks, J.L.; Haffner, M.C.; Rosenberg, A.Z.; Levi, M.; Wang, X.X.; Ozbek, B.; Baena-Del, V.J.; et al. An in Situ Atlas of Mitochondrial DNA in Mammalian Tissues Reveals High Content in Stem and Proliferative Compartments. Am. J. Pathol. 2020, 190, 1565–1579. [Google Scholar] [CrossRef]
- Fukunaga, H. Mitochondrial DNA Copy Number and Developmental Origins of Health and Disease (DOHaD). Int. J. Mol. Sci. 2021, 22, 6634. [Google Scholar] [CrossRef]
- Liu, Q.; Krishnasamy, Y.; Rehman, H.; Lemasters, J.J.; Schnellmann, R.G.; Zhong, Z. Disrupted Renal Mitochondrial Homeostasis after Liver Transplantation in Rats. PLoS ONE 2015, 10, e140906. [Google Scholar] [CrossRef]
- Liao, X.; Lv, X.; Zhang, Y.; Han, Y.; Li, J.; Zeng, J.; Tang, D.; Meng, J.; Yuan, X.; Peng, Z.; et al. Fluorofenidone Inhibits UUO/IRI-Induced Renal Fibrosis by Reducing Mitochondrial Da.amage. Oxid Med. Cell Longev. 2022, 2022, 2453617. [Google Scholar] [CrossRef]
- Fu, Z.J.; Wang, Z.Y.; Xu, L.; Chen, X.H.; Li, X.X.; Liao, W.T.; Ma, H.K.; Jiang, M.D.; Xu, T.T.; Xu, J.; et al. HIF-1alpha-BNIP3-mediated mitophagy in tubular cells protects against renal ischemia/reperfusion injury. Redox Biol 2020, 36, 101671. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Li, L.; Demello, C.; Guo, D.; Jaber, B.L.; Pereira, B.J.; Balakrishnan, V.S. Mitochondrial DNA injury and mortality in hemodialysis patients. J. Am. Soc. Nephrol. 2009, 20, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Siragy, H.M. Pro-renin receptor suppresses mitochondrial biogenesis and function via AMPK/SIRT-1/ PGC-1alpha pathway in diabetic kidney. PLoS ONE 2019, 14, e225728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Chen, Z.; Ma, Y.; Yang, X.; Zhu, Z.; Zhang, Z.; Hu, J.; Liang, W.; Ding, G. AKAP1 contributes to impaired mtDNA replication and mitochondrial dysfunction in podocytes of diabetic kidney disease. Int. J. Biol Sci. 2022, 10, 4026–4042. [Google Scholar] [CrossRef]
- Kaneko, S.; Usui, J.; Hagiwara, M.; Shimizu, T.; Ishii, R.; Takahashi-Kobayashi, M.; Kageyama, M.; Nakada, K.; Hayashi, J.I.; Yamagata, K. Mitochondrial DNA deletion-dependent podocyte injuries in Mito-miceDelta, a murine model of mitochondrial disease. Exp. Anim. 2022, 71, 14–21. [Google Scholar] [CrossRef]
- Xiao, Y.; Clima, R.; Busch, J.; Rabien, A.; Kilic, E.; Villegas, S.L.; Timmermann, B.; Attimonelli, M.; Jung, K.; Meierhofer, D. Decreased Mitochondrial DNA Content Drives OXPHOS Dysregulation in Chromophobe Renal Cell Carcinoma. Cancer Res. 2020, 80, 3830–3840. [Google Scholar] [CrossRef]
- Milenkovic, D.; Sanz-Moreno, A.; Calzada-Wack, J.; Rathkolb, B.; Veronica, A.O.; Gerlini, R.; Aguilar-Pimentel, A.; Misic, J.; Simard, M.L.; Wolf, E.; et al. Mice lacking the mitochondrial exonuclease MGME1 develop inflammatory kidney disease with glomerular dysfunction. PLoS Genet. 2022, 18, e1010190. [Google Scholar] [CrossRef]
- Samuels, D.C.; Li, C.; Li, B.; Song, Z.; Torstenson, E.; Boyd, C.H.; Rokas, A.; Thornton-Wells, T.A.; Moore, J.H.; Hughes, T.M.; et al. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 2013, 9, e1003929. [Google Scholar] [CrossRef] [Green Version]
- Cai, M.; Yu, Q.; Bao, J. A case report of mitochondrial myopathy with membranous nephropathy. BMC Nephrol. 2022, 23, 87. [Google Scholar] [CrossRef]
- Fervenza, F.C.; Gavrilova, R.H.; Nasr, S.H.; Irazabal, M.V.; Nath, K.A. CKD Due to a Novel Mitochondrial DNA Mutation: A Case Report. Am. J. Kidney Dis. 2019, 73, 273–277. [Google Scholar] [CrossRef]
- Lorenz, R.; Ahting, U.; Betzler, C.; Heimering, S.; Borggrafe, I.; Lange-Sperandio, B. Homoplasmy of the Mitochondrial DNA Mutation m.616T>C Leads to Mitochondrial Tubulointerstitial Kidney Disease and Encephalopathia. Nephron 2020, 144, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Bakis, H.; Trimouille, A.; Vermorel, A.; Redonnet, I.; Goizet, C.; Boulestreau, R.; Lacombe, D.; Combe, C.; Martin-Negrier, M.L.; Rigothier, C. Adult onset tubulo-interstitial nephropathy in MT-ND5-related phenotypes. Clin. Genet. 2020, 97, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Morales, N.; Lopez-Domenech, S.; Iannantuoni, F.; Lopez-Gallardo, E.; Sola, E.; Morillas, C.; Rocha, M.; Ruiz-Pesini, E.; Victor, V.M. Mitochondrial DNA Haplogroup JT is Related to Impaired Glycaemic Control and Renal Function in Type 2 Diabetic Patients. J. Clin. Med. 2018, 7, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayr, J.A.; Meierhofer, D.; Zimmermann, F.; Feichtinger, R.; Kogler, C.; Ratschek, M.; Schmeller, N.; Sperl, W.; Kofler, B. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res. 2008, 14, 2270–2275. [Google Scholar] [CrossRef]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Imasawa, T.; Hirano, D.; Nozu, K.; Kitamura, H.; Hattori, M.; Sugiyama, H.; Sato, H.; Murayama, K. Clinicopathologic Features of Mitochondrial Nephropathy. Kidney Int. Rep. 2022, 7, 580–590. [Google Scholar] [CrossRef]
- Bargagli, M.; Primiano, G.; Primiano, A.; Gervasoni, J.; Naticchia, A.; Servidei, S.; Gambaro, G.; Ferraro, P.M. Recurrent kidney stones in a family with a mitochondrial disorder due to the m.3243A>G mutation. Urolithiasis 2019, 47, 489–492. [Google Scholar] [CrossRef]
- De Luise, M.; Guarnieri, V.; Ceccarelli, C.; D’Agruma, L.; Porcelli, A.M.; Gasparre, G. A Nonsense Mitochondrial DNA Mutation Associates with Dysfunction of HIF1alpha in a Von Hippel-Lindau Renal Oncocytoma. Oxid Med. Cell Longev. 2019, 2019, 8069583. [Google Scholar] [CrossRef]
- Lemoine, S.; Panaye, M.; Rabeyrin, M.; Errazuriz-Cerda, E.; Mousson, D.C.B.; Petiot, P.; Juillard, L.; Guebre-Egziabher, F. Renal Involvement in Neuropathy, Ataxia, Retinitis Pigmentosa (NARP) Syndrome: A Case Report. Am. J. Kidney Dis. 2018, 71, 754–757. [Google Scholar] [CrossRef]
- Narumi, K.; Mishima, E.; Akiyama, Y.; Matsuhashi, T.; Nakamichi, T.; Kisu, K.; Nishiyama, S.; Ikenouchi, H.; Kikuchi, A.; Izumi, R.; et al. Focal Segmental Glomerulosclerosis Associated with Chronic Progressive External Ophthalmoplegia and Mitochondrial DNA A3243G Mutation. Nephron 2018, 138, 243–248. [Google Scholar] [CrossRef]
- Connor, T.M.; Hoer, S.; Mallett, A.; Gale, D.P.; Gomez-Duran, A.; Posse, V.; Antrobus, R.; Moreno, P.; Sciacovelli, M.; Frezza, C.; et al. Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet. 2017, 13, e1006620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adema, A.Y.; Janssen, M.C.; van der Heijden, J.W. A novel mutation in mitochondrial DNA in a patient with diabetes, deafness and proteinuria. Neth. J. Med. 2016, 74, 455–457. [Google Scholar] [PubMed]
- Ng, Y.S.; Hardy, S.A.; Shrier, V.; Quaghebeur, G.; Mole, D.R.; Daniels, M.J.; Downes, S.M.; Freebody, J.; Fratter, C.; Hofer, M.; et al. Clinical features of the pathogenic m.5540G>A mitochondrial transfer RNA tryptophan gene mutation. Neuromuscul. Disord. 2016, 26, 702–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabebi, M.; Mkaouar-Rebai, E.; Mnif, M.; Kallabi, F.; Ben, M.A.; Ben, S.W.; Charfi, N.; Keskes-Ammar, L.; Kamoun, H.; Abid, M.; et al. A novel mutation MT-COIII m.9267G>C and MT-COI m.5913G>A mutation in mitochondrial genes in a Tunisian family with maternally inherited diabetes and deafness (MIDD) associated with severe nephropathy. Biochem. Biophys. Res. Commun. 2015, 459, 353–360. [Google Scholar] [CrossRef]
- Imasawa, T.; Tanaka, M.; Yamaguchi, Y.; Nakazato, T.; Kitamura, H.; Nishimura, M. 7501 T > A mitochondrial DNA variant in a patient with glomerulosclerosis. Ren Fail. 2014, 36, 1461–1465. [Google Scholar] [CrossRef] [Green Version]
- Seidowsky, A.; Hoffmann, M.; Glowacki, F.; Dhaenens, C.M.; Devaux, J.P.; de Sainte, F.C.; Provot, F.; Gheerbrant, J.D.; Hummel, A.; Hazzan, M.; et al. Renal involvement in MELAS syndrome—A series of 5 cases and review of the literature. Clin. Nephrol. 2013, 80, 456–463. [Google Scholar] [CrossRef]
- Chan, W.; Ham, Y.H. Probing the Hidden Role of Mitochondrial DNA Damage and Dysfunction in the Etiology of Aristolochic Acid Nephropathy. Chem. Res. Toxicol. 2021, 34, 1903–1909. [Google Scholar] [CrossRef]
- Brinckmann, A.; Weiss, C.; Wilbert, F.; von Moers, A.; Zwirner, A.; Stoltenburg-Didinger, G.; Wilichowski, E.; Schuelke, M. Regionalized pathology correlates with augmentation of mtDNA copy numbers in a patient with myoclonic epilepsy with ragged-red fibers (MERRF-syndrome). PLoS ONE 2010, 5, e13513. [Google Scholar] [CrossRef]
- Jakupciak, J.P.; Maragh, S.; Markowitz, M.E.; Greenberg, A.K.; Hoque, M.O.; Maitra, A.; Barker, P.E.; Wagner, P.D.; Rom, W.N.; Srivastava, S.; et al. Performance of mitochondrial DNA mutations detecting early stage cancer. BMC Cancer 2008, 8, 285. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, H.; Inoue, T.; Ouchi, H.; Jao, T.M.; Inoue, R.; Nishi, H.; Fujii, R.; Ishidate, F.; Tanaka, T.; Tanaka, Y.; et al. Mitochondrial Damage Causes Inflammation via cGAS-STING Signaling in Acute Kidney Injury. Cell Rep. 2019, 29, 1261–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Imam, A.A.; Tombo, N.; Draeger, D.; Bopassa, J.C. RIP3 Translocation into Mitochondria Promotes Mitofilin Degradation to Increase Inflammation and Kidney Injury after Renal Ischemia-Reperfusion. Cells-Basel 2022, 11, 1894. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.W.; Dhillon, P.; Huang, S.; Sheng, X.; Shrestha, R.; Qiu, C.; Kaufman, B.A.; Park, J.; Pei, L.; Baur, J.; et al. Mitochondrial Damage and Activation of the STING Pathway Lead to Renal Inflammation and Fibrosis. Cell Metab. 2019, 30, 784–799. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Nakashima, M.; Ishikiriyama, T.; Nakashima, H.; Yamagata, A.; Imakiire, T.; Kinoshita, M.; Seki, S.; Kumagai, H.; Oshima, N. Effects of L-Carnitine Treatment on Kidney Mitochondria and Macrophages in Mice with Diabetic Nephropathy. Kidney Blood Press Res. 2022, 47, 277–290. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, J.; Peng, F.; Wang, J. Hypermethylation of mitochondrial DNA facilitates bone metastasis of renal cell carcinoma. J. Cancer 2022, 13, 304–312. [Google Scholar] [CrossRef]
- Myakala, K.; Jones, B.A.; Wang, X.X.; Levi, M. Sacubitril/valsartan treatment has differential effects in modulating diabetic kidney disease in db/db mice and KKAy mice compared with valsartan treatment. Am. J. Physiol Ren. Physiol. 2021, 320, F1133–F1151. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Li, C.; Yu, L.; Chang, Y.; Qu, M. Delivery of coenzyme Q10 with mitochondria-targeted nanocarrier attenuates renal ischemia-reperfusion injury in mice. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 131, 112536. [Google Scholar] [CrossRef]
- Ding, M.; Tolbert, E.; Birkenbach, M.; Gohh, R.; Akhlaghi, F.; Ghonem, N.S. Treprostinil reduces mitochondrial injury during rat renal ischemia-reperfusion injury. Biomed. Pharmacother. 2021, 141, 111912. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, R.; Yuan, J.; Da, J.; Zha, Y.; Long, Y. Roxadustat (FG-4592) protects against ischaemia/reperfusion-induced acute kidney injury through inhibiting the mitochondrial damage pathway in mice. Clin. Exp. Pharm. Physiol. 2022, 49, 311–318. [Google Scholar] [CrossRef]
- Yu, X.; Meng, X.; Xu, M.; Zhang, X.; Zhang, Y.; Ding, G.; Huang, S.; Zhang, A.; Jia, Z. Celastrol ameliorates cisplatin nephrotoxicity by inhibiting NF-kappaB and improving mitochondrial function. Ebiomedicine 2018, 36, 266–280. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.; Lu, L.; Zhou, Y.; Liu, J.; Ma, H.; Fu, L.; Huang, S.; Zhang, Y.; Zhang, A.; Jia, Z. The novel STING antagonist H151 ameliorates cisplatin-induced acute kidney injury and mitochondrial dysfunction. Am. J. Physiol. Ren. Physiol. 2021, 320, F608–F616. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; Liu, Z.; He, L. Adiponectin promotes repair of renal tubular epithelial cells by regulating mitochondrial biogenesis and function. Metabolism 2022, 128, 154959. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Li, P.; Luo, Y.; Wu, C.; Liu, Y.; Qin, X.; Huang, X.; Sun, C. Salidroside stimulates the Sirt1/PGC-1alpha axis and ameliorates diabetic nephropathy in mice. Phytomedicine 2019, 54, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Cai, Y.; Wang, Y.; Weng, W.; Chen, Y.; Wang, M.; Zhan, H.; Yu, X.; Wang, T.; Shao, M.; et al. Artemether ameliorates kidney injury by restoring redox imbalance and improving mitochondrial function in Adriamycin nephropathy in mice. Sci. Rep. 2021, 11, 1266. [Google Scholar] [CrossRef]
- Zorova, L.D.; Kovalchuk, S.I.; Popkov, V.A.; Chernikov, V.P.; Zharikova, A.A.; Khutornenko, A.A.; Zorov, S.D.; Plokhikh, K.S.; Zinovkin, R.A.; Evtushenko, E.A.; et al. Do Extracellular Vesicles Derived from Mesenchymal Stem Cells Contain Functional Mitochondria? Int. J. Mol. Sci 2022, 23, 7408. [Google Scholar] [CrossRef]
- Olsen, G.M.; Rinder, H.M.; Tormey, C.A. De novo acquired hemophilia as an immune dysregulation phenomenon following SARS-CoV-2 infection. Transfusion 2021, 61, 989–991. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Y.; Qi, Z.; Cao, L.; Ding, S. Mitochondrial transfer/transplantation: An emerging therapeutic approach for multiple diseases. Cell Biosci. 2022, 12, 66. [Google Scholar] [CrossRef]
- Hernandez-Cruz, E.Y.; Amador-Martinez, I.; Aranda-Rivera, A.K.; Cruz-Gregorio, A.; Pedraza, C.J. Renal damage induced by cadmium and its possible therapy by mitochondrial transplantation. Chem. Biol. Interact. 2022, 361, 109961. [Google Scholar] [CrossRef]
- Fan, X.Y.; Guo, L.; Chen, L.N.; Yin, S.; Wen, J.; Li, S.; Ma, J.Y.; Jing, T.; Jiang, M.X.; Sun, X.H.; et al. Reduction of mtDNA heteroplasmy in mitochondrial replacement therapy by inducing forced mitophagy. Nat. Biomed. Eng. 2022, 6, 339–350. [Google Scholar] [CrossRef]


| Mutation | Gene | Clinical Characters | Diagnosis | Reference |
|---|---|---|---|---|
| m.616T>C | MT-tRNAPhe | Recurrent swelling and pain | CKD | [28] |
| Renal insufficiency | Hyperuricemia | |||
| m.3243A>G | MT-TL1 | Chest tightness and shortness of breath | Membranous nephropathy | [99] |
| Hyperlactatemia | ||||
| Hyperuricemia | ||||
| Proteinuria | ||||
| m.6145G>A | MT-CO1 | Chronic tubulointerstitial changes | Rhabdomyolysis | [100] |
| Elevated serum creatinine level Dark-colored urine Mitochondrial abnormalities | ||||
| m.616T>C | MT-tRNAPhe | Developmental delay Epilepsy Hypertension Electrolyte disturbance Chronic renal insufficiency | Autosomal-dominant tubulointerstitial kidney disease | [101] |
| m.13513G>A m.13514G>A | MT-ND5 | Anuric AKI | Tubulo-interstitial kidney disease | [102] |
| Acute pulmonary edema | ||||
| Hyperlactatemia with metabolic acidosis | ||||
| Proteinuria | ||||
| Hypertension | ||||
| m.4216T>C | MT-ND1 | Higher levels of fasting glucose | DKD | [103] |
| Decreased renal function | ||||
| 3571_3572insC | MT-ND1 | - | Renal oncocytoma | [104] |
| 3571delC | ||||
| 10952_10953insC | MT-ND4 | |||
| 11038delA | ||||
| 12384_12385insT | MT-ND5 | |||
| 12390_12391insC | ||||
| m.13493T>C | ||||
| m.3243A>G | MT-TL1 | |||
| m.3565T>AC | MT-ND1 | - | ChRCC | [105] |
| m.3922G>A | ||||
| m.4569G>A | MT-ND2 | |||
| m.4969G>C | ||||
| m.10806G>A | MT-ND4 | |||
| m.11866A>AC | ||||
| m.12384TC>T | MT-ND5 | |||
| m.12417C>CA | ||||
| m.13127AC>A | ||||
| m.13206CTG>C | ||||
| m.13230CA>C | ||||
| m.14159C>A | MT-ND6 | |||
| m.6490T>C | MT-CO1 | |||
| m.9651C>T | MT-CO3 | |||
| m.3243A>G | MT-TL1 | Proteinuria Decreased eGFR Hyperuricemia | FSGS | [106] |
| Nephrosclerosis | ||||
| DKD | ||||
| Tubulointerstitial nephropathy | ||||
| Minor glomerular abnormality | ||||
| m.3243A>G | MT-TL1 | Osteoporosis | Nephrolithiasis | [107] |
| Bilateral sensorineural deafness | ||||
| Sensory axonal neuropathy | ||||
| m.6129G>A | MT-CO1 | - | Von Hippel-Lindau renal oncocytoma | [108] |
| m.8993T>G | MT-ATP6 | Proteinuria Decreased eGFR | Neuropathy, ataxia and retinitis pigmentosa syndrome | [109] |
| End-stage renal disease | ||||
| m.3243A>G | MT-TL1 | Wolff-Parkinson-White syndrome Proteinuria | Chronic progressive external ophthalmoplegia | [110] |
| FSGS | ||||
| m.547A>T | MT-HSP | Interstitial fibrosis | Tubulointerstitial kidney disease | [111] |
| m.616T>C | MT-tRNAPhe | Tubular atrophy | ||
| m.09155A>G | MT-ATP6 | Central obesity Proteinuria Impaired glucose tolerance | Maternally inherited deafness and diabetes FSGS | [112] |
| m.5540G>A | MT-TW | Proteinuria Hypertension | Cataract | [113] |
| Basal ganglia calcification | ||||
| Retinitis pigmentosa | ||||
| m.9267G>C m.5913G>A | MT-CO3 MT-CO1 | Hypertension | Mitochondrial diabetes DKD | [114] |
| Nephropathy | ||||
| Hyperglycemia | ||||
| Insulin resistance | ||||
| Deafness | ||||
| m.7501T>A | MT-tRNASer | Proteinuria | Glomerulosclerosis Diabetes mellitus | [115] |
| Hypertension | ||||
| Hyperglycemia |
| Therapeutic Interventions | Models | Main Effects on mtDNA and Mitochondrial Function | Reference |
|---|---|---|---|
| Fluorofenidone | UUO and IRI | Increased mtDNA copy number Increased TFAM and PGC-1α expression Maintained mitochondrial structure Reduced mitochondrial oxidative stress | [90] |
| l-carnitine | DKD | Decreased circulating mtDNA content Reduced mtROS production Suppressed inflammation | [124] |
| Sacubitril/valsartan | DKD | Albuminuria Inhibited cGAS-STING signaling Decreased oxidative response | [126] |
| Coenzyme Q10 | IRI | Alleviated mtDNA damage Suppressed inflammatory and oxidative responses | [127] |
| Treprostinil | IRI | Increased mtDNA copy number Increased PGC-1α expression | [128] |
| Increased ATP level | |||
| Reduced mitochondrial oxidative injury | |||
| Roxadustat | IRI | Increased ATPβ and PPARγ expression, and mtDNA | [129] |
| Alleviated DNA damage | |||
| Celastrol | Cisplatin-induced AKI | Increased mtDNA copy number Increased MMP Restored OXPHOS activity | [130] |
| H151 | Cisplatin-induced AKI | Restored mtDNA content Reversed mitochondrial gene expression Suppressed inflammation | [131] |
| Adiponectin | DKD | Increased mtDNA content Increased TFAM and PGC-1α expression Increased mitochondrial mass Increased MMP | [132] |
| Salidroside | DKD | Increased mtDNA copy number Enhanced ETC proteins Increased PGC-1α expression | [133] |
| Artemether | Adriamycin nephropathy | Restored redox imbalance Increased mtDNA copy number | [134] |
| Improved mitochondrial function |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, J.; Chen, Z.; Liang, W.; Wei, Z.; Ding, G. Roles of Mitochondrial DNA Damage in Kidney Diseases: A New Biomarker. Int. J. Mol. Sci. 2022, 23, 15166. https://doi.org/10.3390/ijms232315166
Feng J, Chen Z, Liang W, Wei Z, Ding G. Roles of Mitochondrial DNA Damage in Kidney Diseases: A New Biomarker. International Journal of Molecular Sciences. 2022; 23(23):15166. https://doi.org/10.3390/ijms232315166
Chicago/Turabian StyleFeng, Jun, Zhaowei Chen, Wei Liang, Zhongping Wei, and Guohua Ding. 2022. "Roles of Mitochondrial DNA Damage in Kidney Diseases: A New Biomarker" International Journal of Molecular Sciences 23, no. 23: 15166. https://doi.org/10.3390/ijms232315166