Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use
Abstract
:1. Introduction
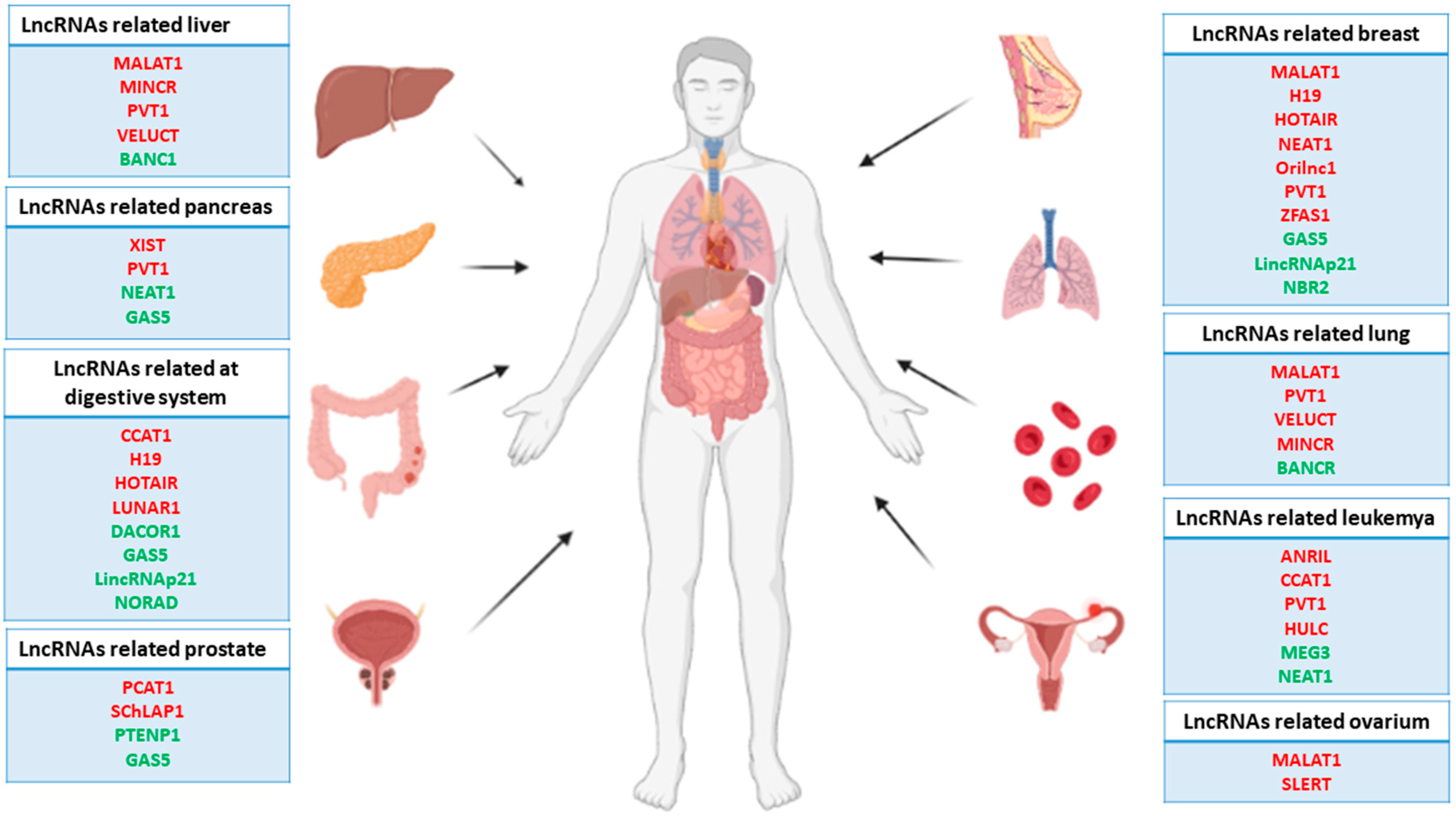
2. Genetic and Epigenetic Contributions of lncRNA Dysregulation in Cancer
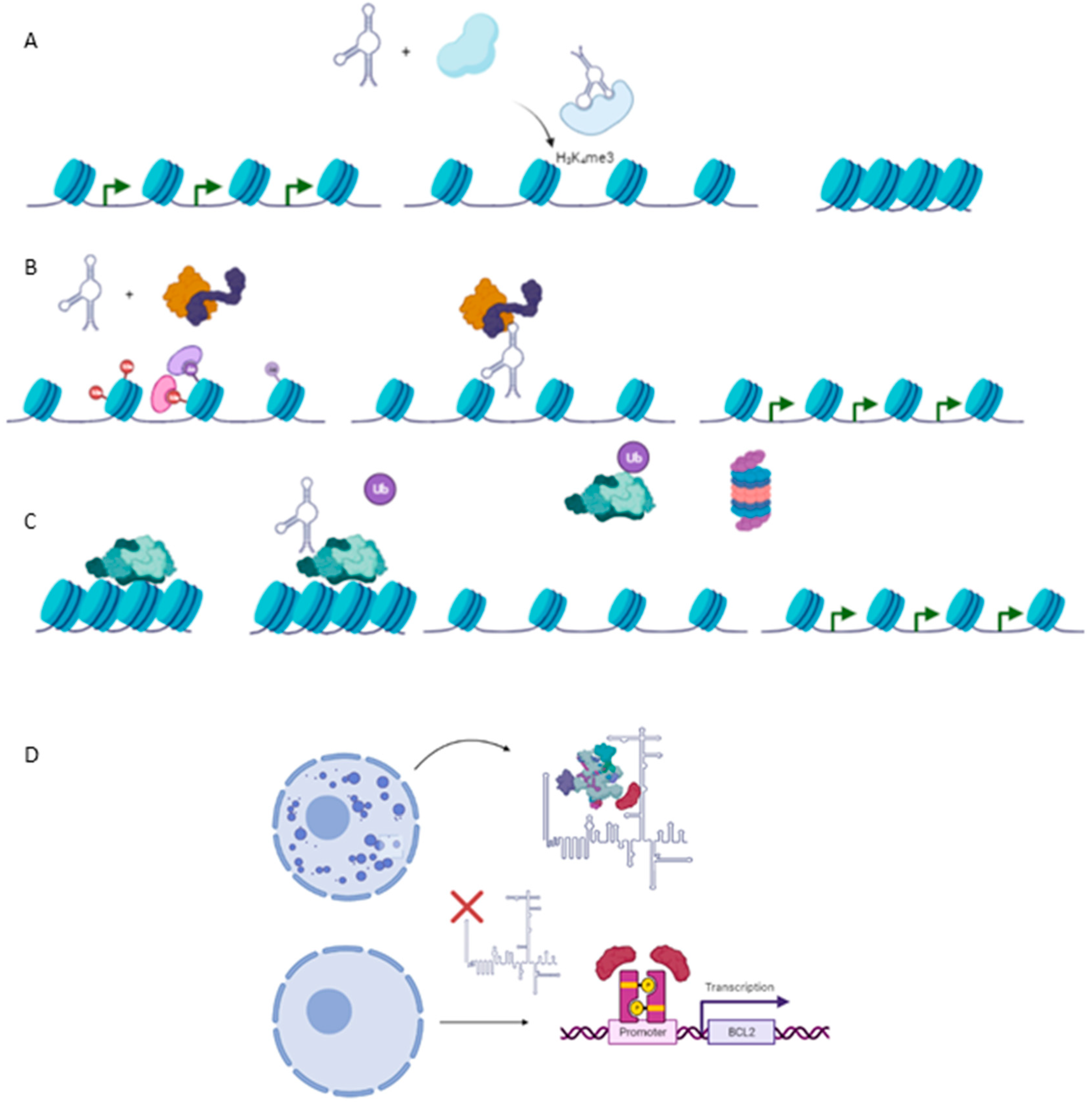
2.1. lncRNAs as Modulators of Histone Methylation
2.2. lncRNAs as Modulators of Histone Acetylation
2.3. lncRNAs as Modulators of DNA Methylation
2.4. lncRNAs as Post-Transcriptional Regulators of Related Epigenetic Proteins
2.5. lncRNAs as Nuclear Environment Modulators
3. Transcriptional Gene Modulation by lncRNAs
4. Post-Transcriptional Gene Regulation by lncRNAs Involved in Cancer
4.1. lncRNAs Acting as Competing Endogenous RNAs
4.2. lncRNAs Promoting or Inhibiting the Translation of mRNA-Targets
4.3. lncRNAs Interact with Ribosome Machinery, Modulating the Biogenesis, Translocation, and Binding of Ribosomal Subunits
4.4. lncRNAs Involved in Splicing Alternative Mechanisms
5. Development of Strategies to Obtain the Blocking Oncogene Functions of lncRNAs
5.1. Genomic Modulation of lncRNAs by a CRISPR-Based System Edition
5.2. Post-Transcriptional Modulation of lncRNAs by Inhibitory Molecules
5.3. Small Molecules against lncRNAs as Therapeutic Drugs
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Gonzalez, D.; et al. The GENCODE v7 Catalogue of Human Long Non-Coding RNAs: Analysis of Their Structure, Evolution and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Hoon, M.; Shin, J.W.; Carninci, P. Paradigm shifts in genomics through the FANTOM projects. Mamm. Genome 2015, 26, 391–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramilowski, J.A.; Yip, C.W.; Agrawal, S.; Chang, J.-C.; Ciani, Y.; Kulakovskiy, I.V.; Mendez, M.; Ooi, J.L.C.; Ouyang, J.F.; Parkinson, N.; et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020, 30, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Haines, J.E.; Perez, E.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef]
- Guh, C.-Y.; Hsieh, Y.-H.; Chu, H.-P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 1–14. [Google Scholar] [CrossRef]
- Chen, Q.W.; Zhu, X.Y.; Li, Y.Y.; Meng, Z.Q. Epigenetic regulation and cancer (Review). Oncol. Rep. 2013, 31, 523–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [Green Version]
- Yin, Q.-F.; Yang, L.; Zhang, Y.; Xiang, J.-F.; Wu, Y.-W.; Carmichael, G.G.; Chen, L.-L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Ounzain, S.; Burdet, F.; Ibberson, M.; Pedrazzini, T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J. Mol. Cell. Cardiol. 2015, 89, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Ounzain, S.; Pedrazzini, T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc. Med. 2015, 25, 592–602. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium; Campbell, P.J.; Getz, G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Ng, A.W.T.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Li, H.; Chen, Q.; Wu, W.; Chen, X.; Ran, L.; Si, G.; Tan, X. A Novel and Robust Long Noncoding RNA Panel to Predict the Prognosis of Pancreatic Cancer. DNA Cell Biol. 2020, 39, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Fu, Y.; Colonna, V.; Mu, X.J.; Kang, H.M.; Lappalainen, T.; Sboner, A.; Lochovsky, L.; Chen, J.; Harmanci, A.; et al. Integrative Annotation of Variants from 1092 Humans: Application to Cancer Genomics. Science 2013, 342, 1235587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unfried, J.P.; Sangro, P.; Prats-Mari, L.; Sangro, B.; Fortes, P. The Landscape of lncRNAs in Hepatocellular Carcinoma: A Translational Perspective. Cancers 2021, 13, 2651. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Chai, H.; Zhao, H. Detecting lncRNA–Cancer Associations by Combining miRNAs, Genes, and Prognosis with Matrix Factorization. Front. Genet. 2021, 12, 639872. [Google Scholar] [CrossRef]
- Vancura, A.; Lanzós, A.; Bosch-Guiteras, N.; Esteban, M.T.; Gutierrez, A.H.; Haefliger, S.; Johnson, R. Cancer LncRNA Census 2 (CLC2): An enhanced resource reveals clinical features of cancer lncRNAs. NAR Cancer 2021, 3, zcab013. [Google Scholar] [CrossRef]
- Ganini, C.; Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Cipriani, C.; Di Daniele, N.; Juhl, H.; Mauriello, A.; et al. Global mapping of cancers: The Cancer Genome Atlas and beyond. Mol. Oncol. 2021, 15, 2823–2840. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Xiu, B.; Chi, Y.; Liu, L.; Chi, W.; Zhang, Q.; Chen, J.; Guo, R.; Si, J.; Li, L.; Xue, J.; et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 2019, 18, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, Y.-S.; Wang, D.-L.; Yang, B.; Yan, H.-Y.; Lin, L.-H.; Li, Y.; Chen, J.; Xie, L.-M.; Liao, J.-Y.; et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 2020, 10, 10823–10837. [Google Scholar] [CrossRef]
- Bian, D.; Gao, C.; Bao, K.; Song, G. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-ĸB. Am. J. Cancer Res. 2017, 7, 28. [Google Scholar]
- Vahidi, S.; Norollahi, S.E.; Agah, S.; Samadani, A.A. DNA Methylation Profiling of hTERT Gene Alongside with the Telomere Performance in Gastric Adenocarcinoma. J. Gastrointest. Cancer 2020, 51, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045–41055. [Google Scholar] [CrossRef] [PubMed]
- Fardi, M.; Solali, S.; Hagh, M.F. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis. 2018, 5, 304–311. [Google Scholar] [CrossRef]
- Richter, A.M.; Woods, M.L.; Küster, M.M.; Walesch, S.K.; Braun, T.; Boettger, T.; Dammann, R.H. RASSF10 is frequently epigenetically inactivated in kidney cancer and its knockout promotes neoplasia in cancer prone mice. Oncogene 2020, 39, 3114–3127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Zhou, M.; Li, X.; Gu, S.; Cao, Y.; Xing, T.; Chen, W.; Chu, C.; Gu, F.; Zhou, J.; et al. SLC34A2 simultaneously promotes papillary thyroid carcinoma growth and invasion through distinct mechanisms. Oncogene 2020, 39, 2658–2675. [Google Scholar] [CrossRef]
- Holleran, J.L.; Parise, R.A.; Joseph, E.; Eiseman, J.L.; Covey, J.M.; Glaze, E.R.; Lyubimov, A.V.; Chen, Y.-F.; D’Argenio, D.Z.; Egorin, M.J. Plasma Pharmacokinetics, Oral Bioavailability, and Interspecies Scaling of the DNA Methyltransferase Inhibitor, Zebularine. Clin. Cancer Res. 2005, 11, 3862–3868. [Google Scholar] [CrossRef] [Green Version]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef] [Green Version]
- Scelfo, A.; Fachinetti, D. Keeping the Centromere under Control: A Promising Role for DNA Methylation. Cells 2019, 8, 912. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Feng, J.; Wang, W.; Deng, Z.; Zhang, Y.; Xiao, L.; Wang, Z.; Liu, C.; Liu, Q.; Chen, S.; et al. The EGFR-ZNF263 signaling axis silences SIX3 in glioblastoma epigenetically. Oncogene 2020, 39, 3163–3178. [Google Scholar] [CrossRef]
- Mirmohammadsadegh, A.; Marini, A.; Nambiar, S.; Hassan, M.; Tannapfel, A.; Ruzicka, T.; Hengge, U.R. Epigenetic Silencing of the PTEN Gene in Melanoma. Cancer Res. 2006, 66, 6546–6552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marini, A.; Mirmohammadsadegh, A.; Nambiar, S.; Gustrau, A.; Ruzicka, T.; Hengge, U.R. Epigenetic Inactivation of Tumor Suppressor Genes in Serum of Patients with Cutaneous Melanoma. J. Investig. Dermatol. 2006, 126, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [Green Version]
- Magaña-Acosta, M.; Valadez-Graham, V. Chromatin Remodelers in the 3D Nuclear Compartment. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Sastry, N.G.; Wan, X.; Huang, T.; Alvarez, A.A.; Pangeni, R.P.; Song, X.; James, C.D.; Horbinski, C.M.; Brennan, C.W.; Nakano, I. LY6K promotes glioblastoma Tumorigenicity via CAV-1-mediated ERK1/2 signaling enhancement. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wu, G.; Zhou, Z.; Zhang, X.; Wang, Y.; Song, G.; Ding, E.; Sun, X.; Zhong, L.; Li, S.; et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial–mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, M.; Mihara, K.; Yasunaga, S.; Ohtaki, M.; Takihara, Y.; Kimura, A. Expression of Polycomb-group (PcG) protein BMI-1 predicts prognosis in patients with acute myeloid leukemia. Leukemia 2007, 6, 846–847. [Google Scholar] [CrossRef] [Green Version]
- Frangini, A.; Sjöberg, M.; Roman-Trufero, M.; Dharmalingam, G.; Haberle, V.; Bartke, T.; Lenhard, B.; Malumbres, M.; Vidal, M.; Dillon, N. The Aurora B Kinase and the Polycomb Protein Ring1B Combine to Regulate Active Promoters in Quiescent Lymphocytes. Mol. Cell 2013, 51, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Barbour, H.; Daou, S.; Hendzel, M.; Affar, E.B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsushima, K.; Natsume, A.; Ohka, F.; Shinjo, K.; Hatanaka, A.; Ichimura, N.; Sato, S.; Takahashi, S.; Kimura, H.; Totoki, Y.; et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016, 7, 13616. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Gao, Z. Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 8594. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, W.; Shao, Z. Association between long non-coding RNA polymorphisms and cancer risk: A meta-analysis. Biosci. Rep. 2018, 38, BSR20180365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, N.; Liu, G.; Pan, Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 2020, 16, 2981–2995. [Google Scholar] [CrossRef]
- Maruei-Milan, R.; Heidari, Z.; Aryan, A.; Asadi-Tarani, M.; Salimi, S. Long non-coding RNA ANRIL polymorphisms in papillary thyroid cancer and its severity. Br. J. Biomed. Sci. 2021, 78, 58–62. [Google Scholar] [CrossRef]
- Zhao, R.; Choi, B.Y.; Lee, M.-H.; Bode, A.M.; Dong, Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16 INK4a) in Cancer. EBioMedicine 2016, 8, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhang, J.-Q.; Chen, J.-Z.; Chen, H.-X.; Qiu, F.-N.; Yan, M.-L.; Chen, Y.-L.; Peng, C.-H.; Tian, Y.-F.; Wang, Y.-D. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: An in vivo and in vitro study. Int. J. Biol. Macromol. 2017, 102, 718–728. [Google Scholar] [CrossRef]
- Wang, C.-H.; Li, Q.-Y.; Nie, L.; Ma, J.; Yao, C.-J.; Chen, F.-P. LncRNA ANRIL promotes cell proliferation, migration and invasion during acute myeloid leukemia pathogenesis via negatively regulating miR-34a. Int. J. Biochem. Cell Biol. 2020, 119, 105666. [Google Scholar] [CrossRef]
- Wang, K.; Hu, Y.-B.; Zhao, Y.; Ye, C. LncRNA ANRIL Regulates Ovarian Cancer Progression and Tumor Stem Cell-Like Characteristics via miR-324-5p/Ran Axis. OncoTargets Ther. 2021, 14, 565–576. [Google Scholar] [CrossRef]
- Sun, L.-Y.; Li, X.-J.; Sun, Y.-M.; Huang, W.; Fang, K.; Han, C.; Chen, Z.-H.; Luo, X.-Q.; Chen, Y.-Q.; Wang, W.-T. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT. Mol. Cancer 2018, 17, 1–6. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Jutooru, I.; Chadalapaka, G.; Corton, J.C.; Safe, S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015, 6, 10840–10852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.; Liu, J.; Guo, S.; He, S.; Qiu, G.; Lu, J.; Wang, J.; Fan, L.; Zhao, W.; Che, X. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol. Rep. 2016, 35, 3577–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Ji, F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol. Lett. 2019, 18, 1649–1656. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Gao, G.; Wang, Z.; Sun, D.; Wei, X.; Ma, Y.; Ding, Y. Long non-coding RNA HOTTIP promotes prostate cancer cells proliferation and migration by sponging miR-216a-5p. Biosci. Rep. 2018, 38, BSR20180566. [Google Scholar] [CrossRef] [Green Version]
- Navarro, A.; Moises, J.; Santasusagna, S.; Marrades, R.M.; Viñolas, N.; Castellano, J.J.; Canals, J.; Muñoz, C.; Ramírez, J.; Molins, L.; et al. Clinical significance of long non-coding RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm. Med. 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Ni, J.; Li, W.; Pan, B.; Yang, Y.; Xia, Q.; Huang, J. The Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade promotes malignant and metastatic progression of osteosarcoma. Mol. Oncol. 2020, 14, 2678–2695. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, H.; Yu, H.; Bi, M.; Yan, Z.; Hong, S.; Li, S. The Long Non-coding RNA HOTTIP Is Highly Expressed in Colorectal Cancer and Enhances Cell Proliferation and Invasion. Mol. Ther. Nucleic Acids 2020, 19, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Oehme, F.; Krahl, S.; Gyorffy, B.; Muessle, B.; Rao, V.; Greif, H.; Ziegler, N.; Lin, K.; Thepkaysone, M.-L.; Polster, H.; et al. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019, 16, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 2015, 6, 1–7, Erratum in Nat. Commun. 2019, 10, 5290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef] [Green Version]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano, R.; Castro, E.; Aragón, I.M.; Cendón, Y.; Cattrini, C.; López-Casas, P.P.; Olmos, D. Genetic aberrations in DNA repair pathways: A cornerstone of precision oncology in prostate cancer. Br. J. Cancer 2021, 124, 552–563. [Google Scholar] [CrossRef]
- Han, J.; Lachance, C.; Ricketts, M.D.; McCullough, C.E.; Gerace, M.; Black, B.E.; Cote, J.; Marmorstein, R. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3–H4 substrate. J. Biol. Chem. 2018, 293, 4498–4509. [Google Scholar] [CrossRef] [Green Version]
- Iizuka, M.; Susa, T.; Takahashi, Y.; Tamamori-Adachi, M.; Kajitani, T.; Okinaga, H.; Fukusato, T.; Okazaki, T. Histone acetyltransferase Hbo1 destabilizes estrogen receptor α by ubiquitination and modulates proliferation of breast cancers. Cancer Sci. 2013, 104, 1647–1655. [Google Scholar] [CrossRef]
- Quintela, M.; Sieglaff, D.H.; Gazze, A.S.; Zhang, A.; Gonzalez, D.; Francis, L.; Webb, P.; Conlan, R.S. HBO1 directs histone H4 specific acetylation, potentiating mechano-transduction pathways and membrane elasticity in ovarian cancer cells. Nanomed. Nanotechnol. Biol. Med. 2019, 17, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Xu, F.; Wang, H.; Teschendorff, A.E.; Xie, F.; He, Y. Pan-cancer characterization of long non-coding RNA and DNA methylation mediated transcriptional dysregulation. EBioMedicine 2021, 68, 103399. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, G.; Mao, P.; Zhang, J.; Zhang, L.; Liu, L.; Wang, J.; Owusu, L.; Ren, B.; Tang, Y.; et al. Down-regulation of GADD45A enhances chemosensitivity in melanoma. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Park, Y.J.; Lindroth, A.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long Noncoding RNA TARID Directs Demethylation and Activation of the Tumor Suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, K.K. DNMT1 as a therapeutic target in pancreatic cancer: Mechanisms and clinical implications. Cell. Oncol. 2020, 43, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Merry, C.R.; Forrest, M.; Sabers, J.N.; Beard, L.; Gao, X.-H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, S.; Forrest, M.E.; Moinova, H.; Cohen, A.; Varadan, V.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. The DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA methylation in colon cancer. Clin. Epigenet. 2018, 10, 127. [Google Scholar] [CrossRef]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, N.; Li, C.; Wang, F. Silencing of LncRNA-ANCR Promotes the Osteogenesis of Osteoblast Cells in Postmenopausal Osteoporosis via Targeting EZH2 and RUNX2. Yonsei Med. J. 2019, 60, 751–759. [Google Scholar] [CrossRef]
- Wen, Z.; Lian, L.; Ding, H.; Hu, Y.; Xiao, Z.; Xiong, K.; Yang, Q. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020, 17, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Furth, N.; Aylon, Y. The LATS1 and LATS2 tumor suppressors: Beyond the Hippo pathway. Cell Death Differ. 2017, 24, 1488–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Cai, Q.; Wang, S.; Wang, S.; Mondal, T.; Wang, J.; Quan, Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.-H.; You, B.-H.; Park, C.H.; Kim, Y.J.; Nam, J.-W.; Kil Lee, S. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018, 417, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmehdawi, F.; Wheway, G.; Szymanska, K.; Adams, M.; High, A.S.; Johnson, C.A.; Robinson, P.A. Human Homolog of Drosophila Ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp. Cell Res. 2013, 319, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Krejci, J.; Legartová, S.; Bártová, E. Neural Differentiation in HDAC1-Depleted Cells Is Accompanied by Coilin Downregulation and the Accumulation of Cajal Bodies in Nucleoli. Stem. Cells Int. 2017, 2017, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Shastrula, P.K.; Sierra, I.; Deng, Z.; Keeney, F.; Hayden, J.E.; Lieberman, P.M.; Janicki, S.M. PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. J. Cell Sci. 2019, 132, jcs.220970. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Lei, Y.-Y.; Ding, M.-G.; Luo, L.-H.; Xie, Y.-C.; Liu, X.-L. LncRNA NEAT1 Interacted with DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front. Genet. 2020, 11, 250. [Google Scholar] [CrossRef]
- Zhu, Y.; Tomlinson, R.L.; Lukowiak, A.A.; Terns, R.M.; Terns, M.P. Telomerase RNA Accumulates in Cajal Bodies in Human Cancer Cells. Mol. Biol. Cell 2004, 15, 81–90. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, J.; Sampieri, K.; Clohessy, J.G.; Mendez, L.; Gonzalez-Billalabeitia, E.; Liu, X.-S.; Lee, Y.-R.; Fung, J.; Katon, J.M.; et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 2018, 50, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, Q.; Wang, Q.; Shen, Q.; Chen, X.; Li, Z.; Zhou, Y.; Hou, J.; Xu, B.; Li, N.; et al. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. OncoImmunology 2018, 7, e1503913. [Google Scholar] [CrossRef] [Green Version]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Liu, S.; Lu, S.; Yu, X.; Lai, J.; Wu, Y.; Chen, S.; Wang, L.; Yu, Z.; Luo, G.; et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 2018, 17, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating Gene Expression through RNA Nuclear Retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef] [Green Version]
- Ben-Zvi, M.; Amariglio, N.; Paret, G.; Nevo-Caspi, Y. F11R Expression upon Hypoxia Is Regulated by RNA Editing. PLoS ONE 2013, 8, e77702. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Shao, C.; Wu, Q.-J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.-T.; Zhang, Y.; Wang, Y.; et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017, 24, 816–824. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Harvey, A.R.; Hodgetts, S.I.; Fox, A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 2017, 23, 872–881. [Google Scholar] [CrossRef] [Green Version]
- West, J.A.; Mito, M.; Kurosaka, S.; Takumi, T.; Tanegashima, C.; Chujo, T.; Yanaka, K.; Kingston, R.E.; Hirose, T.; Bond, C.; et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016, 214, 817–830. [Google Scholar] [CrossRef]
- Rowley, J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef]
- Savic, D.; Roberts, B.S.; Carleton, J.B.; Partridge, E.C.; White, M.A.; Cohen, B.A.; Cooper, G.M.; Gertz, J.; Myers, R.M. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/enhancer-binding protein beta binding sites. Genome Res. 2015, 25, 1791–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spicuglia, S.; Vanhille, L. Chromatin signatures of active enhancers. Nucleus 2012, 3, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Chng, W.-J.; Zhou, J. Super-enhancers: Critical roles and therapeutic targets in hematologic malignancies. J. Hematol. Oncol. 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2020, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Adeel, M.M.; Jiang, H.; Arega, Y.; Cao, K.; Lin, D.; Cao, C.; Cao, G.; Wu, P.; Li, G. Structural Variations of the 3D Genome Architecture in Cervical Cancer Development. Front. Cell Dev. Biol. 2021, 9, 706375. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liang, H. A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super-enhancer Activities in Cancer. Cancer Cell 2020, 38, 701–715. [Google Scholar] [CrossRef]
- Deng, R.; Huang, J.-H.; Wang, Y.; Zhou, L.-H.; Wang, Z.-F.; Hu, B.-X.; Chen, Y.-H.; Yang, D.; Mai, J.; Li, Z.-L.; et al. Disruption of super-enhancer-driven tumor suppressor gene RCAN1.4 expression promotes the malignancy of breast carcinoma. Mol. Cancer 2020, 19, 1–19. [Google Scholar] [CrossRef]
- Lidschreiber, K.; Jung, L.A.; von der Emde, H.; Dave, K.; Taipale, J.; Cramer, P.; Lidschreiber, M. Transcriptionally active enhancers in human cancer cells. Mol. Syst. Biol. 2021, 17, e9873. [Google Scholar] [CrossRef]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef] [Green Version]
- Rosati, E.; Baldoni, S.; De Falco, F.; Del Papa, B.; Dorillo, E.; Rompietti, C.; Albi, E.; Falzetti, F.; Di Ianni, M.; Sportoletti, P. NOTCH1 Aberrations in Chronic Lymphocytic Leukemia. Front. Oncol. 2018, 8, 229. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Deng, Y.; Yan, X.; Zhu, J.; Yin, Y.; Shu, Y.; Bai, D.; Zhang, S.; Xu, H.; Lu, X. The Role of ARID5B in Acute Lymphoblastic Leukemia and Beyond. Front. Genet. 2020, 11, 598. [Google Scholar] [CrossRef]
- Tan, S.H.; Leong, W.Z.; Ngoc, P.C.T.; Tan, T.K.; Bertulfo, F.C.; Lim, M.C.; An, O.; Li, Z.; Yeoh, A.E.J.; Fullwood, M.J.; et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood 2019, 134, 239–251. [Google Scholar] [CrossRef]
- Shi, X.; Sun, M.; Wu, Y.; Yao, Y.; Liu, H.; Wu, G.; Yuan, D.; Song, Y. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumor Biol. 2015, 36, 503–513. [Google Scholar] [CrossRef]
- Zhang, G.; Li, H.; Sun, R.; Li, P.; Yang, Z.; Liu, Y.; Wang, Z.; Yang, Y.; Yin, C. Long non-coding RNA ZEB2-AS1 promotes the proliferation, metastasis and epithelial mesenchymal transition in triple-negative breast cancer by epigenetically activating ZEB2. J. Cell. Mol. Med. 2019, 23, 3271–3279. [Google Scholar] [CrossRef] [Green Version]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A.; Babbs, B.; Cochrane, D.R.; Bitler, B.G.; Richer, J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol. Carcinog. 2019, 58, 196–205. [Google Scholar] [CrossRef]
- Lan, Y.; Xiao, X.; He, Z. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in col-orectal cancer. Nucleic Acids Res. 2018, 46, 5809–5821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Wu, J.; Wang, J.Y. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degra-dation of MicroRNA 195. Gastroenterology 2018, 154, 599–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karreth, F.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.; Rust, A.; DeNicola, G.; Webster, K.; Weiss, D.; Perez-Mancera, P.A.; et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell 2011, 147, 382–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poliseno, L.; Haimovic, A.; Christos, P.J.; de Miera, E.C.V.Y.S.; Shapiro, R.; Pavlick, A.; Berman, R.; Darvishian, F.; Osman, I. Deletion of PTENP1 Pseudogene in Human Melanoma. J. Investig. Dermatol. 2011, 131, 2497–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Ren, W.; Zhang, L.; Li, S.; Kong, X.; Zhang, H.; Dong, J.; Cai, G.; Jin, C.; Zheng, D.; et al. PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 2017, 56, 1322–1334. [Google Scholar] [CrossRef]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells through an miR-9–Mediated RXRA Signaling Pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of Highly Up-regulated in Liver Cancer (HULC) by Hepatitis B Virus X Protein Promotes Hepatoma Cell Proliferation via Down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Li, Y.; Chai, X.; Kang, Q.; Zhao, P.; Xiong, J.; Wang, J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene 2017, 607, 41–46. [Google Scholar] [CrossRef]
- Xin, X.; Wu, M.; Meng, Q.; Wang, C.; Lu, Y.; Yang, Y.; Li, X.; Zheng, Q.; Pu, H.; Gui, X.; et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Xiao, Z.; Liu, F.; Cui, M.; Li, W.; Yang, Z.; Li, J.; Ye, L.; Zhang, X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget 2016, 7, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wu, T.; Zhou, H.; Jin, Q.; He, G.; Yu, H.; Xuan, L.; Wang, X.; Tian, L.; Sun, Y.; et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J. Exp. Clin. Cancer Res. 2016, 35, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xiao, Z.; Du, X.; Huang, L.; Du, G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol. Rep. 2016, 37, 555–562. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-H.; Zhang, S.-Q.; Qiu, X.-G.; Zhang, S.-J.; Zheng, S.-H.; Zhang, D.-H. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int. J. Oncol. 2017, 50, 708–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhao, X.; Guo, Z.; Ma, X.; Song, Y.; Guo, Y. Regulation of NEAT1/miR-214-3p on the growth, migration and invasion of endometrial carcinoma cells. Arch. Gynecol. Obstet. 2017, 295, 1469–1475. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, Y.; Wang, N.; Yu, N. NEAT1 negatively regulates miR-218 expression and promotes breast cancer progression. Cancer Biomark. 2017, 20, 247–254. [Google Scholar] [CrossRef]
- Qian, K.; Liu, G.; Tang, Z.; Hu, Y.; Fang, Y.; Chen, Z.; Xu, X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2017, 615, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.; Li, Z.; Long, X.; Guo, Z.; Zhang, G.; Zu, J.; Chen, Y.; Wen, L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int. J. Biol. Macromol. 2017, 105, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Liu, C.; Wu, Q.; Zhang, J.; Min, Q.; Sheng, T.; Wang, X.; Zou, Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem. Biophys. Res. Commun. 2017, 482, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lei, Z.-J.; Guo, Y.; Wang, T.; Qin, Z.-Y.; Xiao, H.-L.; Fan, L.-L.; Chen, D.-F.; Bian, X.-W.; Liu, J.; et al. miRNA-regulated delivery of lincRNA-p21 suppresses β-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget 2015, 6, 37852–37870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Habiba, U.; Kuroshima, T.; Yanagawa-Matsuda, A.; Kitamura, T.; Chowdhury, A.; Jehung, J.P.; Hossain, E.; Sano, H.; Kitagawa, Y.; Shindoh, M.; et al. HuR translocation to the cytoplasm of cancer cells in actin-independent manner. Exp. Cell Res. 2018, 369, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.; Gorospe, M. LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 2012, 47, 648–655, Erratum in Mol. Cell 2013, 50, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.-H.; Yao, R.-W.; Zhang, Y.; Guo, C.-J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.-L. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 2017, 169, 664–678.e16. [Google Scholar] [CrossRef]
- Lindqvist, L.M.; Tandoc, K.; Topisirovic, I.; Furic, L. Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr. Opin. Genet. Dev. 2018, 48, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Xu, G.; Han, C.; Luan, P.-F.; Xing, Y.-H.; Nan, F.; Yang, L.-Z.; Huang, Y.; Yang, Z.-H.; Shan, L.; et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 2021, 373, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Álvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [Green Version]
- Lan, T.; Chang, L.; Wu, L.; Yuan, Y. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol. Med. Rep. 2016, 14, 4606–4612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tang, L. The Application of lncRNAs in Cancer Treatment and Diagnosis. Recent Pat. Anti-Cancer Drug Discov. 2018, 13, 292–301. [Google Scholar] [CrossRef]
- Hofmans, M.; Lammens, T.; Depreter, B.; Wu, Y.; Erlacher, M.; Caye, A.; Cavé, H.; Flotho, C.; de Haas, V.; Niemeyer, C.M.; et al. Long non-coding RNAs as novel therapeutic targets in juvenile myelomonocytic leukemia. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Mahboobeh, Z.; Pegah, M.; Fatemeh, S.; Elham, K.; Hanieh, A.; Milad, R.; Mohammad, S. lncRNA ZEB2-AS1: A promising biomarker in human cancers. IUBMB Life 2020, 72, 1891–1899. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Myacheva, K.; Groß, M.; Klingenberg, M.; Arqué, B.D.; Diederichs, S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2016, 45, e12. [Google Scholar] [CrossRef] [Green Version]
- Aparicio-Prat, E.; Arnan, C.; Sala, I.; Bosch, N.; Guigó, R.; Johnson, R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genom. 2015, 16, 1–15, Erratum in BMC Genom. 2016, 17, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toden, S.; Zumwalt, T.J.; Goel, A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta 2021, 1875, 188491. [Google Scholar] [CrossRef]
- Koch, L. Functional genomics: Screening for lncRNA function. Nat. Rev. Genet. 2017, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, X.; Yuan, J.; Geng, T.; Chen, S.; Hu, X.; Cui, I.H.; Cui, H. Biallelic insertion of a transcriptional terminator via the CRISPR/Cas9 system efficiently silences expression of protein-coding and non-coding RNA genes. J. Biol. Chem. 2017, 292, 5624–5633. [Google Scholar] [CrossRef] [Green Version]
- Santos, R.D.S.; Pinheiro, D.P.; Teixeira, L.P.R.; Sales, S.L.A.; Luciano, M.C.D.S.; Melo, M.M.D.L.; Pinheiro, R.F.; Tavares, K.C.S.; Furtado, G.P.; Pessoa, C.; et al. CRISPR/Cas9 small promoter deletion in H19 lncRNA is associated with altered cell morphology and proliferation. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Pauli, A.; Montague, T.G.; Lennox, K.A.; Behlke, M.; Schier, A.F. Antisense Oligonucleotide-Mediated Transcript Knockdown in Zebrafish. PLoS ONE 2015, 10, e0139504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T.A.; Zou, L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 2017, 65, 832–847. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, Y.; Jin, J.; Han, G.; Sun, C.; Pizzi, M.P.; Huo, L.; Scott, A.; Wang, Y.; Ma, L.; et al. LncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol. Cancer 2019, 18, 1–5, Erratum in Mol. Cancer 2021, 20, 56. [Google Scholar] [CrossRef]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Wheeler, T.M.; Leger, A.J.; Pandey, S.K.; MacLeod, A.R.; Nakamori, M.; Cheng, S.H.; Wentworth, B.M.; Bennett, C.F.; Thornton, C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nat. Cell Biol. 2012, 488, 111–115. [Google Scholar] [CrossRef] [Green Version]
- Gong, N.; Teng, X.; Li, J.; Liang, X.-J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef]
- Kurreck, J. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [Green Version]
- Sarma, K.; Levasseur, P.; Aristarkhov, A.; Lee, J.T. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA 2010, 107, 22196–22201. [Google Scholar] [CrossRef] [Green Version]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.; Fatemi, R.P.; Magistri, M.; Brothers, S.; Van Der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-Stranded Antisense siRNAs Guide Target RNA Cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Prensner, J.; Iyer, M.K.; Sahu, A.; Asangani, I.; Cao, Q.; Patel, L.; Vergara, I.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taxman, D.J.; Moore, C.B.; Guthrie, E.H.; Huang, M.T.-H. Short Hairpin RNA (shRNA): Design, Delivery, and Assessment of Gene Knockdown. In Methods in Molecular Biology; Springer: Heidelberg, Germany, 2010; Volume 629, pp. 139–156. [Google Scholar]
- Sun, R.; Qin, C.; Jiang, B.; Fang, S.; Pan, X.; Peng, L.; Liu, Z.; Li, W.; Li, Y.; Li, G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial–mesenchymal transition. Mol. BioSyst. 2016, 12, 952–962. [Google Scholar] [CrossRef]
- Zong, Z.; Song, Y.; Xue, Y.; Ruan, X.; Liu, X.; Yang, C.; Zheng, J.; Cao, S.; Li, Z.; Liu, Y. Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviours of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway. J. Cell. Mol. Med. 2019, 23, 5048–5062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.; Arun, G.; Nam, H.; Schneekloth, J.S.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Padilla, C.; Dueñas, Á.; García-López, V.; Aránega, A.; Franco, D.; Garcia-Martínez, V.; López-Sánchez, C. Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use. Int. J. Mol. Sci. 2022, 23, 764. https://doi.org/10.3390/ijms23020764
García-Padilla C, Dueñas Á, García-López V, Aránega A, Franco D, Garcia-Martínez V, López-Sánchez C. Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use. International Journal of Molecular Sciences. 2022; 23(2):764. https://doi.org/10.3390/ijms23020764
Chicago/Turabian StyleGarcía-Padilla, Carlos, Ángel Dueñas, Virginio García-López, Amelia Aránega, Diego Franco, Virginio Garcia-Martínez, and Carmen López-Sánchez. 2022. "Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use" International Journal of Molecular Sciences 23, no. 2: 764. https://doi.org/10.3390/ijms23020764





