Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients: A Preliminary Study
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. MicroRNA Profiling in Plasma of Normal and Breast Cancer Patients
2.2.1. Phase 1: Pilot Testing of miRNA Biomarkers
2.2.2. Phase 2: Final Testing
2.3. Identification of Deregulated miRNA by the ddCt Method
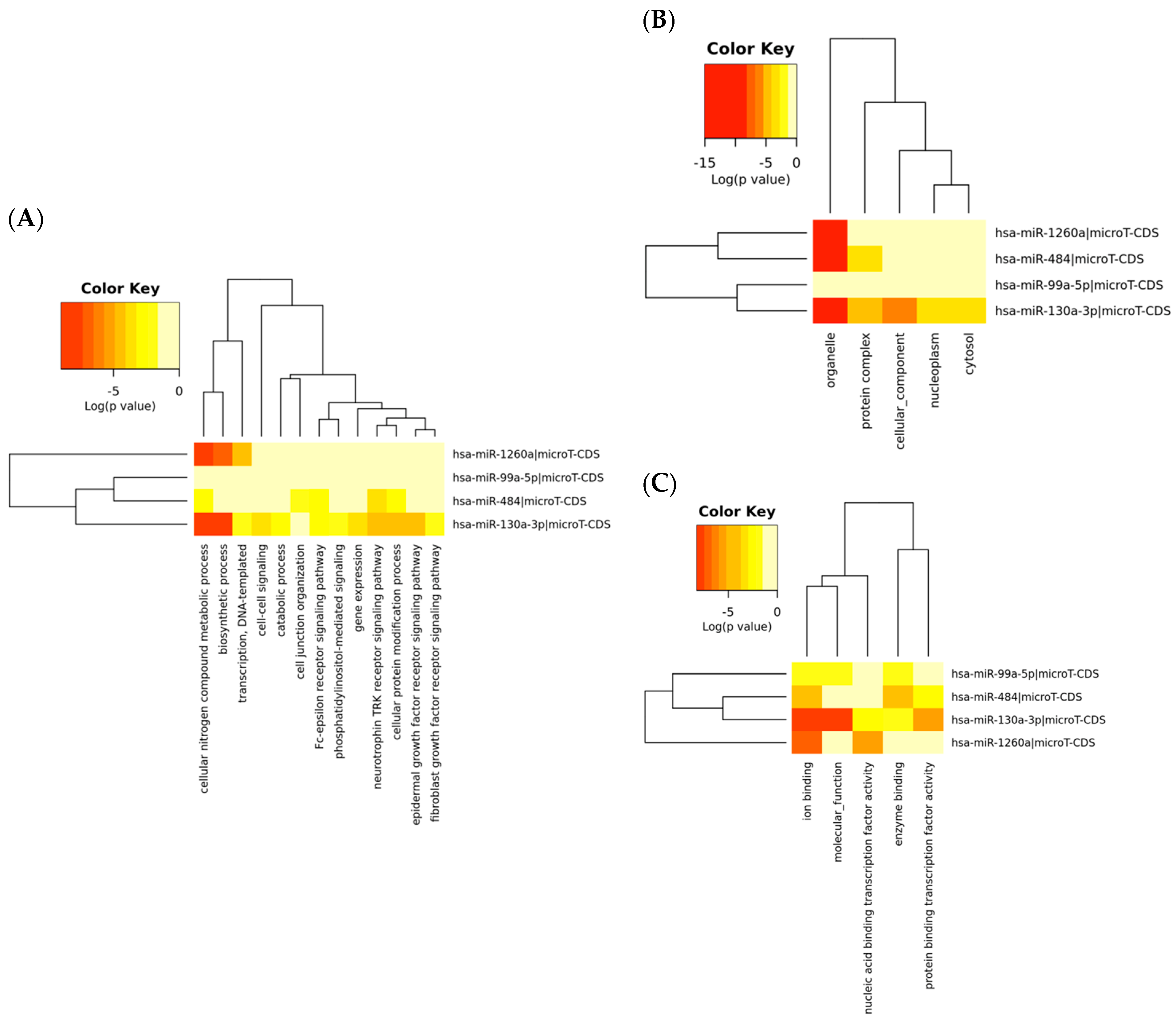
2.4. Signal and Functional Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment, Blood Collection and Plasma Preparation
4.2. RNA Extraction, Reverse Transcription of RNA and Quality Control
4.3. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
4.4. Statistical Analysis and Bioinformatics
4.5. Signalling and Functional Analysis of Differentially Expressed miRNAs
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dafni, U.; Tsourti, Z.; Alatsathianos, I. Breast Cancer Statistics in the European Union: Incidence and Survival across European Countries. Breast Care 2019, 14, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Fuller, M.S.; Lee, C.I.; Elmore, J.G. Breast Cancer Screening: An Evidence-Based Update. Med. Clin. N. Am. 2015, 99, 451–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.-J. Strategies for subtypes—dealing with the diversity of breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Gadzicki, D.; Evans, D.G.; Harris, H.; Julian-Reynier, C.; Nippert, I.; Schmidtke, J.; Tibben, A.; van Asperen, C.J.; Schlegelberger, B. Genetic testing for familial/hereditary breast cancer—Comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J. Community Genet. 2011, 2, 53–69. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, A.; Song, M.; Brook, M.N.; Milne, R.L.; Mavaddat, N.; Michailidou, K.; Bolla, M.K.; Wang, Q.; Dennis, J.; Wilcox, A.N.; et al. Joint associations of a polygenic risk score and environmental risk factors for breast cancer in the Breast Cancer Association Consortium. Int. J. Epidemiol. 2018, 47, 526–536. [Google Scholar] [CrossRef] [Green Version]
- Chang-Claude, J.; Eby, N.; Kiechle, M.; Bastert, G.; Becher, H. Breastfeeding and breast cancer risk by age 50 among women in Germany. Cancer Causes Control CCC 2000, 11, 687–695. [Google Scholar] [CrossRef]
- Horn, J.; Vatten, L.J. Reproductive and hormonal risk factors of breast cancer: A historical perspective. Int. J. Womens Health 2017, 9, 265–272. [Google Scholar] [CrossRef] [Green Version]
- McDonald, K.-A.; Young, J. Breast Cancer Screening Modalities. In Preventive Oncology for the Gynecologist; Mehta, S., Singla, A., Eds.; Springer: Singapore, 2019; pp. 353–366. ISBN 9789811334382. [Google Scholar]
- Zubor, P.; Kubatka, P.; Kajo, K.; Dankova, Z.; Polacek, H.; Bielik, T.; Kudela, E.; Samec, M.; Liskova, A.; Vlcakova, D.; et al. Why the Gold Standard Approach by Mammography Demands Extension by Multiomics? Application of Liquid Biopsy miRNA Profiles to Breast Cancer Disease Management. Int. J. Mol. Sci. 2019, 20, 2878. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.H.; Montano, M.A. Chapter 1—MicroRNAs: Mirrors of Health and Disease. In Translating MicroRNAs to the Clinic; Laurence, J., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 1–15. ISBN 978-0-12-800553-8. [Google Scholar]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a–363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.; Roberts, H.; Rai, S.N.; Galandiuk, S. Housekeeping genes for studies of plasma microRNA: A need for more precise standardization. Surgery 2015, 158, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Wang, Z.; Liao, B.-Y.; Yu, L.; Gao, X.; Lu, S.; Wang, S.; Dai, Z.; Zhang, X.; Chen, Q.; et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int. J. Cancer 2014, 135, 1187–1194. [Google Scholar] [CrossRef]
- Zografos, E.; Zagouri, F.; Kalapanida, D.; Zakopoulou, R.; Kyriazoglou, A.; Apostolidou, K.; Gazouli, M.; Dimopoulos, M.-A. Prognostic role of microRNAs in breast cancer: A systematic review. Oncotarget 2019, 10, 7156–7178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhavan, D.; Peng, C.; Wallwiener, M.; Zucknick, M.; Nees, J.; Schott, S.; Rudolph, A.; Riethdorf, S.; Trumpp, A.; Pantel, K.; et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis 2016, 37, 461–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1–research0034.11. [Google Scholar] [CrossRef] [Green Version]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [Green Version]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy: Potential and challenges. Mol. Oncol. 2016, 10, 371–373. [Google Scholar] [CrossRef] [Green Version]
- Luu-The, V.; Paquet, N.; Calvo, E.; Cumps, J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. BioTechniques 2005, 38, 287–293. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Minicozzi, A.M.; Benati, M.; Paviati, E.; Lima-Oliveira, G.; Gusella, M.; Pasini, F.; Salvagno, G.L.; Montagnana, M.; Lippi, G. Reference miRNAs for colorectal cancer: Analysis and verification of current data. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, V.Y.; Siu, J.M.; Cheuk, I.; Ng, E.K.O.; Kwong, A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.K.O.; Li, R.; Shin, V.Y.; Jin, H.C.; Leung, C.P.H.; Ma, E.S.K.; Pang, R.; Chua, D.; Chu, K.-M.; Law, W.L.; et al. Circulating microRNAs as Specific Biomarkers for Breast Cancer Detection. PLoS ONE 2013, 8, e53141. [Google Scholar] [CrossRef] [Green Version]
- Rinnerthaler, G.; Hackl, H.; Gampenrieder, S.P.; Hamacher, F.; Hufnagl, C.; Hauser-Kronberger, C.; Zehentmayr, F.; Fastner, G.; Sedlmayer, F.; Mlineritsch, B.; et al. miR-16-5p Is a Stably-Expressed Housekeeping MicroRNA in Breast Cancer Tissues from Primary Tumors and from Metastatic Sites. Int. J. Mol. Sci. 2016, 17, 156. [Google Scholar] [CrossRef]
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of Circulating miRNAs in Plasma: Effect of Preanalytical and Analytical Parameters on Their Isolation and Stability. J. Mol. Diagn. 2013, 15, 827–834. [Google Scholar] [CrossRef]
- Chan, M.; Liaw, C.S.; Ji, S.M.; Tan, H.H.; Wong, C.Y.; Thike, A.A.; Tan, P.H.; Ho, G.H.; Lee, A.S.-G. Identification of Circulating MicroRNA Signatures for Breast Cancer Detection. Clin. Cancer Res. 2013, 19, 4477–4487. [Google Scholar] [CrossRef] [Green Version]
- Graveel, C.R.; Calderone, H.M.; Westerhuis, J.J.; Winn, M.E.; Sempere, L.F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer Targets Ther. 2015, 7, 59–79. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.T.-H.; Wang, F.; Chapin, W.; Huang, R.S. Identification of MicroRNAs as Breast Cancer Prognosis Markers through the Cancer Genome Atlas. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA Targeting Dicer for Metastasis Control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolacinska, A.; Morawiec, J.; Fendler, W.; Malachowska, B.; Morawiec, Z.; Szemraj, J.; Pawlowska, Z.; Chowdhury, D.; Choi, Y.E.; Kubiak, R.; et al. Association of microRNAs and pathologic response to preoperative chemotherapy in triple negative breast cancer: Preliminary report. Mol. Biol. Rep. 2014, 41, 2851–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.; Quan, H.; Lv, J.; Meng, L.; Wang, C.; Yu, Z.; Han, J. Serum microRNA as potential biomarker to detect breast atypical hyperplasia and early-stage breast cancer. Future Oncol. Lond. Engl. 2018, 14, 3145–3161. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Rosa, A.; Guedes, L.C.; Fonseca, B.V.; Gotovac, K.; Violante, S.; Mestre, T.; Coelho, M.; Rosa, M.M.; Martin, E.R.; et al. Convergence of miRNA Expression Profiling, α-Synuclein Interacton and GWAS in Parkinson’s Disease. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Meyer, H.-A.; Jung, M.; Fendler, A.; Wagner, I.; Stephan, C.; Busch, J.; Erbersdobler, A.; Disch, A.C.; Mollenkopf, H.-J.; et al. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal. Biochem. 2011, 417, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Q.; Tang, L. MiR-99a Antitumor Activity in Human Breast Cancer Cells through Targeting of mTOR Expression. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 2012, 12, 369. [Google Scholar] [CrossRef] [Green Version]
- Oneyama, C.; Ikeda, J.; Okuzaki, D.; Suzuki, K.; Kanou, T.; Shintani, Y.; Morii, E.; Okumura, M.; Aozasa, K.; Okada, M. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene 2011, 30, 3489–3501. [Google Scholar] [CrossRef] [Green Version]
- Long, X.; Shi, Y.; Ye, P.; Guo, J.; Zhou, Q.; Tang, Y. MicroRNA-99a Suppresses Breast Cancer Progression by Targeting FGFR3. Front. Oncol. 2020, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Han, Y.; Cheng, K.; Zhang, G.; Wang, X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014, 47, 587–595. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Zhang, X.; Lin, Y.; Luo, T.; Xiao, Z.; Zhou, Q. A dual PI3K/AKT/mTOR signaling inhibitor miR-99a suppresses endometrial carcinoma. Am. J. Transl. Res. 2016, 8, 719–731. [Google Scholar] [PubMed]
- Yu, X.; Liang, J.; Xu, J.; Li, X.; Xing, S.; Li, H.; Liu, W.; Liu, D.; Xu, J.; Huang, L.; et al. Identification and Validation of Circulating MicroRNA Signatures for Breast Cancer Early Detection Based on Large Scale Tissue-Derived Data. J. Breast Cancer 2018, 21, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Cui, R.; Bahr, J.; Zanesi, N.; Luo, Z.; Meng, W.; Liang, G.; Croce, C.M. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017, 77, 6168–6178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.-L.; Liu, B.; Shi, Y.-F.; Liu, N.; Yan, Y.-Y.; Zhang, J.-C.; Xue, X.; Wang, J.-P.; Zhao, Z.; Liu, J.-G.; et al. MicroRNA-130a alleviates human coronary artery endothelial cell injury and inflammatory responses by targeting PTEN via activating PI3K/Akt/eNOS signaling pathway. Oncotarget 2016, 7, 71922–71936. [Google Scholar] [CrossRef]
- Duan, J.; Zhang, H.; Qu, Y.; Deng, T.; Huang, D.; Liu, R.; Zhang, L.; Bai, M.; Zhou, L.; Ying, G.; et al. Onco-miR-130 promotes cell proliferation and migration by targeting TGFβR2 in gastric cancer. Oncotarget 2016, 7, 44522–44533. [Google Scholar] [CrossRef] [Green Version]
- Stückrath, I.; Rack, B.; Janni, W.; Jäger, B.; Pantel, K.; Schwarzenbach, H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 2015, 6, 13387–13401. [Google Scholar] [CrossRef]
- Braicu, C.; Raduly, L.; Morar-Bolba, G.; Cojocneanu, R.; Jurj, A.; Pop, L.-A.; Pileczki, V.; Ciocan, C.; Moldovan, A.; Irimie, A.; et al. Aberrant miRNAs expressed in HER-2 negative breast cancers patient. J. Exp. Clin. Cancer Res. 2018, 37, 257. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, L.; Sun, D.; Li, J.; Ji, Z. The role of miR-130a in cancer. Breast Cancer 2017, 24, 521–527. [Google Scholar] [CrossRef]
- Zearo, S.; Kim, E.; Zhu, Y.; Zhao, J.T.; Sidhu, S.B.; Robinson, B.G.; Soon, P.S. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer 2014, 14, 200. [Google Scholar] [CrossRef]
- Vecchione, A.; Belletti, B.; Lovat, F.; Volinia, S.; Chiappetta, G.; Giglio, S.; Sonego, M.; Cirombella, R.; Onesti, E.C.; Pellegrini, P.; et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc. Natl. Acad. Sci. USA. 2013, 110, 9845–9850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, Z.; Sun, C.; Gong, H. High serum miR-484 expression is associated with the diagnosis and prognosis of patients with non-small cell lung cancer. Exp. Ther. Med. 2019, 18, 4095–4102. [Google Scholar] [CrossRef] [Green Version]
- Rzepiel, A.; Kutszegi, N.; Gézsi, A.; Sági, J.C.; Egyed, B.; Péter, G.; Butz, H.; Nyírő, G.; Müller, J.; Kovács, G.T.; et al. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef] [PubMed]
- Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Silveyra, P.; Podany, A.; Birch, L.L.; Paul, I.M.; Kelleher, S.; Hicks, S.D. Metabolism-related microRNAs in maternal breast milk are influenced by premature delivery. Pediatr. Res. 2017, 82, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Sugita, B.M.; Pereira, S.R.; de Almeida, R.C.; Gill, M.; Mahajan, A.; Duttargi, A.; Kirolikar, S.; Fadda, P.; de Lima, R.S.; Urban, C.A.; et al. Integrated copy number and miRNA expression analysis in triple negative breast cancer of Latin American patients. Oncotarget 2019, 10, 6184–6203. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Burwinkel, B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012, 9, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- McGuire, A.; Brown, J.A.L.; Kerin, M.J. Metastatic breast cancer: The potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015, 34, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 13 July 2020).
- Eklund, A. Beeswarm: The Bee Swarm Plot, an Alternative to Stripchart version 0.2.3 from CRAN. Available online: https://rdrr.io/cran/beeswarm/ (accessed on 13 July 2020).
- Maechler, M.; Rousseeuw, P.; Croux, C.; Todorov, V.; Ruckstuhl, A.; Salibian-Barrera, M.; Verbeke, T.; Koller, M.; Conceicao, E.; diPalma, M.A. Robustbase: Basic Robust Statistics. R package version 0.93-6. Available online: http://robustbase.r-forge.r-project.org/ (accessed on 13 July 2020).
- Mair, P.; Wilcox, R. Robust statistical methods in R using the WRS2 package. Behav. Res. Methods 2020, 52, 464–488. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Number of Patients (%) | |
|---|---|---|
| All patients | 65 (100%) | |
| Age | Median (IQR) | 56 (47,68) |
| Missing | 5 | |
| Histology | Ductal | 21 (40%) |
| Lobular | 8 (15%) | |
| Invasive cancer, NST | 24 (45%) | |
| Missing | 12 | |
| Subtype | Luminal A | 33 (63%) |
| Luminal B | 13 (25%) | |
| HER2 | 4 (7.7%) | |
| TNC | 2 (3.8%) | |
| Missing | 13 | |
| Grade | 1 | 8 (14%) |
| 2 | 30 (53%) | |
| 3 | 19 (33%) | |
| Missing | 8 | |
| ER status | Positive | 48 (87%) |
| Negative | 7 (13%) | |
| Missing | 10 | |
| PR status | Positive | 43 (78%) |
| Negative | 12 (22%) | |
| Missing | 10 | |
| HER2 | Positive | 9 (18%) |
| Negative | 41 (82%) | |
| Missing | 15 | |
| Tumour size | TIS | 4 (6.9%) |
| <20 mm | 38 (66%) | |
| 20 mm–50 mm | 13 (22%) | |
| >50 mm | 3 (5.2%) | |
| Missing | 7 | |
| Nodal status | Negative | 27 (56%) |
| N1 | 17 (35%) | |
| N2 | 4 (8.3%) | |
| Missing | 17 | |
| LVI | Positive | 28 (51%) |
| Negative | 27 (49%) | |
| Missing | 10 | |
| PNI | Positive | 11 (20%) |
| Negative | 44 (80%) | |
| Missing | 10 | |
| ki67 | ≤15% | 34 (65%) |
| >15% | 18 (35%) | |
| Missing | 13 |
| miRNA ID | GeneGlobe Data Analysis Tool | ddCt Method | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Pilot Testing | Final Testing | ||||||||
| FC | Reg. | p-value | FC | Reg. | p-value | FC | Reg. | p-value | |
| miR-10b | 1.24 | ↑ | 0.17 | 0.86 | ↓ | 0.88 | 0.91 | ↓ | 0.68 |
| miR-16 | 0.95 | ↓ | 0.45 | 0.95 | ↓ | 0.40 | 1 | - | 1 |
| miR-99a | 1.27 | ↑ | 0.22 | 1.06 | ↑ | 0.44 | 1.1 | ↑ | 0.03 |
| miR-103a | 1.06 | ↑ | 0.37 | 1.05 | ↑ | 0.62 | 1 | - | 1 |
| miR-130a | 0.94 | ↓ | 0.91 | 1.01 | ↑ | 0.34 | 1.33 | ↑ | <0.01 |
| miR-191 | 1.18 | ↑ | 0.13 | - | - | - | - | - | - |
| miR-342 | 0.67 | ↓ | 0.05 | 0.76 | ↓ | 0.37 | 0.76 | ↓ | 0.35 |
| miR-484 | 1.35 | ↑ | 0.01 | 1.14 | ↑ | 0.36 | 1.22 | ↑ | <0.01 |
| miR-486 | 0.90 | ↓ | 0.66 | 0.92 | ↓ | 0.34 | 0.89 | ↓ | 0.37 |
| miR-520d | 1.43 | ↑ | 0.31 | - | - | - | - | - | - |
| miR-1228 | 1.43 | ↑ | 0.31 | - | - | - | - | - | - |
| miR-1260a | 0.82 | ↓ | 0.58 | 1.03 | ↑ | 0.12 | 1.2 | ↑ | <0.01 |
| KEGG Pathway | Pathway ID | p-Value | miRNAs | Target Genes |
|---|---|---|---|---|
| Prion diseases | hsa05020 | 3.17 × 10−22 | miR-130a-3p | PNRP |
| Hippo signalling pathway | hsa04390 | 0.007 | miR-130a-3p | WNT2B, FRMD6, TGFB2, TGFBR2, BMPR2, PPP1CB |
| miR-484 | YAP1, WNT2B, WWC1, TP53BP2, CDH1, AXIN2, DLG2, TCF7, FGF1 | |||
| miR-99a-5p | FDZ5, FDZ8, SMAD7 | |||
| miR-1260a | DLG2 | |||
| Phosphatidylinositol signalling system | hsa04070 | 0.0082 | miR-130a-3p | CDS1, DGKE, CALM2, PLCB1, PIKFYVE, PIK3C2A, PTEN, PLCB4, DGKH |
| miR-484 | CALM1, PLCZ1, PIKFYVE, PIK3CD | |||
| miR-1260a | PIP4K2C, DGKH | |||
| Oestrogen signalling pathway | hsa04915 | 0.0136 | miR-484 | CREB3L3, CALM1, PIK3CD, GRM1, KCNJ5 |
| miR-130a-3p | ESR1, ADCY1, SOS2, CALM2, PLCB1, KCNJ6, HSPA8, PLCB4 | |||
| miR-1260a | ADCY1, ATF6B, KCNJ6, SP1 | |||
| Glioma | hsa05214 | 0.0136 | miR-484 | CALM1, PIK3CD, PDGFA |
| miR-130a-3p | SOS2, E2F2, TGFA, CALM2, IGF1, CDKN1A, PTEN | |||
| miR-99a-5p | MTOR | |||
| TGF-beta signalling pathway | hsa04350 | 0.0389 | miR-484 | ACVR1B, PITX2 |
| miR-130a-3p | INHBB, SMURF2, INHBA, ACVR1, SKP1, ZFYVE9, SMAD5, TGFB2, TGFBR2, BMPR2 | |||
| miR-1260a | SKP1, SP1 | |||
| miR-99a-5p | SMAD7 | |||
| Glycosaminoglycan biosynthesis—heparan sulphate/heparin | hsa00534 | 0.0399 | miR-1260a | EXT2, HS3ST2 |
| miR-99a-5p | HS3ST3B1, HS3ST2 | |||
| mTOR signalling pathway | hsa04150 | 0.0399 | miR-99a-5p | MTOR |
| miR-130a-3p | TSC1, RRAGD, PRKAA2, PRKAA1, IGF1, EIF4E2, PTEN, ULK2 | |||
| miR-484 | RPS6KA1, PIK3CD |
| GO Term | p-Value | Target Gene Account |
|---|---|---|
| Biological process | ||
| GO:0034641: cellular nitrogen compound metabolic process | 1.11 × 10−16 | 387 |
| GO:0009058: biosynthetic process | 4.86 × 10−14 | 241 |
| GO:0048011: neurotrophin TRK receptor signalling pathway | 8.09 × 10−6 | 31 |
| GO:0006464: cellular protein modification process | 1.64 × 10−5 | 151 |
| GO:0007173: epidermal growth factor receptor signalling pathway | 0.0002 | 17 |
| GO:0006351: transcription, DNA-templated | 0.0007 | 154 |
| GO:0038095: Fc-epsilon receptor signalling pathway | 0.0015 | 20 |
| GO:0010467: gene expression | 0.0035 | 25 |
| GO:0007267: cell-cell signalling | 0.0188 | 32 |
| GO:0009056: catabolic process | 0.0250 | 64 |
| GO:0048015: phosphatidylinositol-mediated signalling | 0.0315 | 10 |
| GO:0034330: cell junction organisation | 0.0471 | 11 |
| GO:0008543: fibroblast growth factor receptor signalling pathway | 0.0497 | 11 |
| Cellular component | ||
| GO:0043226: organelle | <1 × 10−325 | 772 |
| GO:0043234: protein complex | 3.68 × 10−6 | 239 |
| GO:0005575: cellular component | 0.00018 | 496 |
| GO:0005829: cytosol | 0.00667 | 96 |
| GO:0005654: nucleoplasm | 0.00765 | 47 |
| Molecular function | ||
| GO:0043167: ion binding | <1 × 10−325 | Top of Form 488 Bottom of Form |
| GO:0003674: molecular function | 1.374 × 10−7 | Top of Form 515 Bottom of Form |
| GO:0001071: nucleic acid binding transcription factor activity | 3.503 × 10−6 | Top of Form 74 Bottom of Form |
| GO:0000988: protein binding transcription factor activity | 1.861 × 10−5 | Top of Form 50 Bottom of Form |
| GO:0019899: enzyme binding | 5.208 × 10−5 | Top of Form 98 Bottom of Form |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holubekova, V.; Kolkova, Z.; Grendar, M.; Brany, D.; Dvorska, D.; Stastny, I.; Jagelkova, M.; Zelinova, K.; Samec, M.; Liskova, A.; et al. Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients: A Preliminary Study. Int. J. Mol. Sci. 2020, 21, 7288. https://doi.org/10.3390/ijms21197288
Holubekova V, Kolkova Z, Grendar M, Brany D, Dvorska D, Stastny I, Jagelkova M, Zelinova K, Samec M, Liskova A, et al. Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients: A Preliminary Study. International Journal of Molecular Sciences. 2020; 21(19):7288. https://doi.org/10.3390/ijms21197288
Chicago/Turabian StyleHolubekova, Veronika, Zuzana Kolkova, Marian Grendar, Dusan Brany, Dana Dvorska, Igor Stastny, Marianna Jagelkova, Katarina Zelinova, Marek Samec, Alena Liskova, and et al. 2020. "Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients: A Preliminary Study" International Journal of Molecular Sciences 21, no. 19: 7288. https://doi.org/10.3390/ijms21197288
APA StyleHolubekova, V., Kolkova, Z., Grendar, M., Brany, D., Dvorska, D., Stastny, I., Jagelkova, M., Zelinova, K., Samec, M., Liskova, A., Laucekova, Z., Kudela, E., Bobrovska, M., Kalman, M., Zubor, P., & Dankova, Z. (2020). Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients: A Preliminary Study. International Journal of Molecular Sciences, 21(19), 7288. https://doi.org/10.3390/ijms21197288





