Olfactory-Related Quality of Life in Multiple Chemical Sensitivity: A Genetic-Acquired Factors Model
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Genotype Analysis
4.2.1. Allelic Discrimination by Real-Time PCR
4.2.2. Sanger Sequencing
4.2.3. Multiplex PCR Analysis for Deletion Polymorphisms
4.3. Olfactory Study
4.4. Data Handling and Statistical Analysis
5. Limitations of the Study
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alessandrini, M.; Micarelli, A.; Chiaravalloti, A.; Bruno, E.; Danieli, R.; Pierantozzi, M.; Genovesi, G.; Oberg, J.; Pagani, M.; Schillaci, O. Involvement of Subcortical Brain Structures During Olfactory Stimulation in Multiple Chemical Sensitivity. Brain Topogr. 2016, 29, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Dantoft, T.M.; Elberling, J.; Brix, S.; Szecsi, P.B.; Vesterhauge, S.; Skovbjerg, S. An elevated pro-inflammatory cytokine profile in multiple chemical sensitivity. Psychoneuroendocrinology 2014, 40, 140–150. [Google Scholar] [CrossRef]
- Rossi, S.; Pitidis, A. Multiple Chemical Sensitivity: Review of the State of the Art in Epidemiology, Diagnosis, and Future Perspectives. J. Occup. Environ. Med. 2018, 60, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Das-Munshi, J.; Rubin, G.J.; Wessely, S. Multiple chemical sensitivities: A systematic review of provocation studies. J. Allergy Clin. Immunol. 2006, 118, 1257–1264. [Google Scholar] [CrossRef]
- Karnekull, S.C.; Jonsson, F.U.; Larsson, M.; Olofsson, J.K. Affected by smells? Environmental chemical responsivity predicts odor perception. Chem. Senses 2011, 36, 641–648. [Google Scholar] [CrossRef]
- Viziano, A.; Micarelli, A.; Pasquantonio, G.; Della-Morte, D.; Alessandrini, M. Perspectives on multisensory perception disruption in idiopathic environmental intolerance: A systematic review. Int. Arch. Occup. Environ. Health 2018, 91, 923–935. [Google Scholar] [CrossRef]
- Bascom, R.; Meggs, W.J.; Frampton, M.; Hudnell, K.; Killburn, K.; Kobal, G.; Medinsky, M.; Rea, W. Neurogenic inflammation: With additional discussion of central and perceptual integration of nonneurogenic inflammation. Environ. Health Perspect. 1997, 105 (Suppl. 2), 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winters, W.; Devriese, S.; Van Diest, I.; Nemery, B.; Veulemans, H.; Eelen, P.; Van de Woestijne, K.; Van den Bergh, O. Media warnings about environmental pollution facilitate the acquisition of symptoms in response to chemical substances. Psychosom. Med. 2003, 65, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pall, M.L. Elevated nitric oxide/peroxynitrite theory of multiple chemical sensitivity: Central role of N-methyl-D-aspartate receptors in the sensitivity mechanism. Environ. Health Perspect. 2003, 111, 1461–1464. [Google Scholar] [CrossRef]
- Wiesmuller, G.A.; Niggemann, H.; Weissbach, W.; Riley, F.; Maarouf, Z.; Dott, W.; Kunert, H.J.; Zerres, K.; Eggermann, T.; Blomeke, B. Sequence variations in subjects with self-reported multiple chemical sensitivity (sMCS): A case-control study. J. Toxicol. Environ. Health A 2008, 71, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Bell, I.R.; Brooks, A.J.; Howerter, A.; Jackson, N.; Schwartz, G.E. Acute electroencephalographic effects from repeated olfactory administration of homeopathic remedies in individuals with self-reported chemical sensitivity. Altern. Ther. Health Med. 2013, 19, 46–57. [Google Scholar] [PubMed]
- Alessandrini, M.; Micarelli, A.; Bruno, E.; Ottaviani, F.; Conetta, M.; Cormano, A.; Genovesi, G. Intranasal administration of hyaluronan as a further resource in olfactory performance in multiple chemical sensitivity syndrome. Int. J. Immunopathol. Pharm. 2013, 26, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Chiaravalloti, A.; Pagani, M.; Micarelli, A.; Di Pietro, B.; Genovesi, G.; Alessandrini, M.; Schillaci, O. Cortical activity during olfactory stimulation in multiple chemical sensitivity: A (18)F-FDG PET/CT study. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 733–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papo, D.; Eberlein-Konig, B.; Berresheim, H.W.; Huss-Marp, J.; Grimm, V.; Ring, J.; Behrendt, H.; Winneke, G. Chemosensory function and psychological profile in patients with multiple chemical sensitivity: Comparison with odor-sensitive and asymptomatic controls. J. Psychosom. Res. 2006, 60, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.; Claeson, A.S.; Nyberg, L.; Stenberg, B.; Nordin, S. Brain responses to olfactory and trigeminal exposure in idiopathic environmental illness (IEI) attributed to smells—An fMRI study. J. Psychosom. Res. 2014, 77, 401–408. [Google Scholar] [CrossRef]
- Zhang, J.X.; Liu, Y.J.; Zhang, J.H.; Sun, L. Dual role of preputial gland secretion and its major components in sex recognition of mice. Physiol. Behav. 2008, 95, 388–394. [Google Scholar] [CrossRef]
- Nodari, F.; Hsu, F.F.; Fu, X.; Holekamp, T.F.; Kao, L.F.; Turk, J.; Holy, T.E. Sulfated steroids as natural ligands of mouse pheromone-sensing neurons. J. Neurosci. 2008, 28, 6407–6418. [Google Scholar] [CrossRef]
- Haga, S.; Hattori, T.; Sato, T.; Sato, K.; Matsuda, S.; Kobayakawa, R.; Sakano, H.; Yoshihara, Y.; Kikusui, T.; Touhara, K. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 2010, 466, 118–122. [Google Scholar] [CrossRef]
- Leinders-Zufall, T.; Ishii, T.; Mombaerts, P.; Zufall, F.; Boehm, T. Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat. Neurosci. 2009, 12, 1551–1558. [Google Scholar] [CrossRef]
- Touhara, K. Sexual communication via peptide and protein pheromones. Curr. Opin. Pharm. 2008, 8, 759–764. [Google Scholar] [CrossRef]
- Trotier, D. Vomeronasal organ and human pheromones. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 184–190. [Google Scholar] [CrossRef]
- Rodewald, A.; Mills, D.; Gebhart, V.M.; Jirikowski, G.F. Steroidal pheromones and their potential target sites in the vomeronasal organ. Steroids 2019, 142, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Ploss, V.; Gebhart, V.M.; Dolz, W.; Jirikowski, G.F. Sex hormone binding globulin in the rat olfactory system. J. Chem. Neuroanat. 2014, 57–58, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Ploss, V.M.; Gebhart, V.M.; Gisder, D.; Dolz, W.; Jirikowski, G.F. Localization of sex hormone binding globulin in the rat vomeronasal organ. J. Chem. Neuroanat. 2014, 61–62, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Rodewald, A.; Gebhart, V.M.; Oehring, H.; Jirikowski, G.F. The rat vomeronasal organ is a vitamin D target. J. Chem. Neuroanat. 2017, 81, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Cui, J.; Shen, Y. Brain sex matters: Estrogen in cognition and Alzheimer’s disease. Mol. Cell. Endocrinol. 2014, 389, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcion, E.; Wion-Barbot, N.; Montero-Menei, C.N.; Berger, F.; Wion, D. New clues about vitamin D functions in the nervous system. Trends Endocrinol. Metab. 2002, 13, 100–105. [Google Scholar] [CrossRef]
- Ganji, V.; Milone, C.; Cody, M.M.; McCarty, F.; Wang, Y.T. Serum vitamin D concentrations are related to depression in young adult US population: The Third National Health and Nutrition Examination Survey. Int. Arch. Med. 2010, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Jorde, R.; Waterloo, K.; Saleh, F.; Haug, E.; Svartberg, J. Neuropsychological function in relation to serum parathyroid hormone and serum 25-hydroxyvitamin D levels. The Tromso study. J. Neurol. 2006, 253, 464–470. [Google Scholar] [CrossRef]
- Greene, G.J.; Kipen, H.M. The vomeronasal organ and chemical sensitivity: A hypothesis. Environ. Health Perspect. 2002, 110 (Suppl. 4), 655–661. [Google Scholar] [CrossRef] [Green Version]
- De Luca, C.; Raskovic, D.; Pacifico, V.; Thai, J.C.; Korkina, L. The search for reliable biomarkers of disease in multiple chemical sensitivity and other environmental intolerances. Int. J. Environ. Res. Public Health 2011, 8, 2770–2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labarge, X.S.; McCaffrey, R.J. Multiple chemical sensitivity: A review of the theoretical and research literature. Neuropsychol. Rev. 2000, 10, 183–211. [Google Scholar] [CrossRef] [PubMed]
- Sorg, B.A. Multiple chemical sensitivity: Potential role for neural sensitization. Crit. Rev. Neurobiol. 1999, 13, 283–316. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.; Claeson, A.S.; Dantoft, T.M.; Skovbjerg, S.; Lind, N.; Nordin, S. Chemosensory perception, symptoms and autonomic responses during chemical exposure in multiple chemical sensitivity. Int. Arch. Occup. Environ. Health 2016, 89, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Dantoft, T.M.; Andersson, L.; Nordin, S.; Skovbjerg, S. Chemical intolerance. Curr. Rheumatol. Rev. 2015, 11, 167–184. [Google Scholar] [CrossRef] [Green Version]
- Yunus, M.B. Editorial review: An update on central sensitivity syndromes and the issues of nosology and psychobiology. Curr. Rheumatol. Rev. 2015, 11, 70–85. [Google Scholar] [CrossRef]
- Light, A.R.; Bateman, L.; Jo, D.; Hughen, R.W.; Vanhaitsma, T.A.; White, A.T.; Light, K.C. Gene expression alterations at baseline and following moderate exercise in patients with Chronic Fatigue Syndrome and Fibromyalgia Syndrome. J. Intern. Med. 2012, 271, 64–81. [Google Scholar] [CrossRef]
- Dantoft, T.M.; Skovbjerg, S.; Andersson, L.; Claeson, A.S.; Engkilde, K.; Lind, N.; Nordin, S.; Hellgren, L.I. Gene expression profiling in persons with multiple chemical sensitivity before and after a controlled n-butanol exposure session. BMJ Open 2017, 7, 013879. [Google Scholar] [CrossRef]
- Fujimori, S.; Hiura, M.; Yi, C.X.; Xi, L.; Katoh, T. Factors in genetic susceptibility in a chemical sensitive population using QEESI. Environ. Health Prev. Med. 2012, 17, 357–363. [Google Scholar] [CrossRef] [Green Version]
- McKeown-Eyssen, G.; Baines, C.; Cole, D.E.; Riley, N.; Tyndale, R.F.; Marshall, L.; Jazmaji, V. Case-control study of genotypes in multiple chemical sensitivity: CYP2D6, NAT1, NAT2, PON1, PON2 and MTHFR. Int. J. Epidemiol. 2004, 33, 971–978. [Google Scholar] [CrossRef]
- Schnakenberg, E.; Fabig, K.R.; Stanulla, M.; Strobl, N.; Lustig, M.; Fabig, N.; Schloot, W. A cross-sectional study of self-reported chemical-related sensitivity is associated with gene variants of drug-metabolizing enzymes. Environ. Health 2007, 6, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korkina, L.; Scordo, M.G.; Deeva, I.; Cesareo, E.; De Luca, C. The chemical defensive system in the pathobiology of idiopathic environment-associated diseases. Curr. Drug Metab. 2009, 10, 914–931. [Google Scholar] [CrossRef]
- Caccamo, D.; Cesareo, E.; Mariani, S.; Raskovic, D.; Ientile, R.; Curro, M.; Korkina, L.; De Luca, C. Xenobiotic sensor- and metabolism-related gene variants in environmental sensitivity-related illnesses: A survey on the Italian population. Oxid. Med. Cell. Longev. 2013, 2013, 831969. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Thai, J.C.; Raskovic, D.; Cesareo, E.; Caccamo, D.; Trukhanov, A.; Korkina, L. Metabolic and genetic screening of electromagnetic hypersensitive subjects as a feasible tool for diagnostics and intervention. Mediat. Inflamm. 2014, 2014, 924184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loria-Kohen, V.; Marcos-Pasero, H.; de la Iglesia, R.; Aguilar-Aguilar, E.; Espinosa-Salinas, I.; Herranz, J.; Ramirez de Molina, A.; Reglero, G. Multiple chemical sensitivity: Genotypic characterization, nutritional status and quality of life in 52 patients. Med. Clin. (Barc) 2017, 149, 141–146. [Google Scholar] [CrossRef]
- D’Attis, S.; Massari, S.; Mazzei, F.; Maio, D.; Vergallo, I.; Mauro, S.; Minelli, M.; Bozzetti, M.P. Assessment of CYP2C9, CYP2C19, and CYP2D6 Polymorphisms in Allergic Patients with Chemical Sensitivity. Int. Arch. Allergy Immunol. 2019, 179, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Pall, M.L. Common etiology of posttraumatic stress disorder, fibromyalgia, chronic fatigue syndrome and multiple chemical sensitivity via elevated nitric oxide/peroxynitrite. Med. Hypotheses 2001, 57, 139–145. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Scordo, M.G.; Cesareo, E.; Pastore, S.; Mariani, S.; Maiani, G.; Stancato, A.; Loreti, B.; Valacchi, G.; Lubrano, C.; et al. Biological definition of multiple chemical sensitivity from redox state and cytokine profiling and not from polymorphisms of xenobiotic-metabolizing enzymes. Toxicol. Appl. Pharm. 2010, 248, 285–292. [Google Scholar] [CrossRef]
- Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; Zhu, G.D.; Voland, J.; Moreno, T.A. Frequency of CYP2D6 Alleles Including Structural Variants in the United States. Front. Pharm. 2018, 9, 305. [Google Scholar] [CrossRef] [Green Version]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Klein, T.E.; Caudle, K.E.; Haidar, C.E.; Shen, D.D.; Callaghan, J.T.; Sadhasivam, S.; et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharm. Ther. 2014, 95, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Berg, N.D.; Rasmussen, H.B.; Linneberg, A.; Brasch-Andersen, C.; Fenger, M.; Dirksen, A.; Vesterhauge, S.; Werge, T.; Elberling, J. Genetic susceptibility factors for multiple chemical sensitivity revisited. Int. J. Hyg. Environ. Health 2010, 213, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lu, X.; Hiura, M.; Oda, M.; Miyazaki, W.; Katoh, T. Evaluation of genetic polymorphisms in patients with multiple chemical sensitivity. PLoS ONE 2013, 8, e73708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antognelli, C.; Del Buono, C.; Ludovini, V.; Gori, S.; Talesa, V.N.; Crino, L.; Barberini, F.; Rulli, A. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: An Italian case-control study. BMC Cancer 2009, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, A.; Fassett, R.G.; Geraghty, D.P.; Kunde, D.A.; Ball, M.J.; Robertson, I.K.; Coombes, J.S. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 2012, 501, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Palmirotta, R.; Barbanti, P.; De Marchis, M.L.; Egeo, G.; Aurilia, C.; Fofi, L.; Ialongo, C.; Valente, M.G.; Ferroni, P.; Della-Morte, D.; et al. Is SOD2 Ala16Val polymorphism associated with migraine with aura phenotype? Antioxid. Redox Signal. 2015, 22, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Malinowska, K.; Kowalski, M.; Szaflik, J.; Szaflik, J.P.; Majsterek, I. The role of Cat -262C/T, GPX1 Pro198Leu and Sod1+35A/C gene polymorphisms in a development of primary open-angle glaucoma in a Polish population. Pol. J. Pathol. 2016, 67, 404–410. [Google Scholar] [CrossRef] [Green Version]
- De Luca, C.; Gugliandolo, A.; Calabro, C.; Curro, M.; Ientile, R.; Raskovic, D.; Korkina, L.; Caccamo, D. Role of polymorphisms of inducible nitric oxide synthase and endothelial nitric oxide synthase in idiopathic environmental intolerances. Mediat. Inflamm. 2015, 2015, 245308. [Google Scholar] [CrossRef]
- Serpe, L.; Canaparo, R.; Scordo, M.G.; Spina, E. Pharmacogenetics of drug-metabolizing enzymes in Italian populations. Drug Metab. Pers. Ther. 2015, 30, 107–120. [Google Scholar] [CrossRef]
- Martis, S.; Peter, I.; Hulot, J.S.; Kornreich, R.; Desnick, R.J.; Scott, S.A. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharm. J. 2013, 13, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Serrano, D.; Lazzeroni, M.; Zambon, C.F.; Macis, D.; Maisonneuve, P.; Johansson, H.; Guerrieri-Gonzaga, A.; Plebani, M.; Basso, D.; Gjerde, J.; et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharm. J. 2011, 11, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Boccia, S.; Sayed-Tabatabaei, F.A.; Persiani, R.; Gianfagna, F.; Rausei, S.; Arzani, D.; La Greca, A.; D’Ugo, D.; La Torre, G.; van Duijn, C.M.; et al. Polymorphisms in metabolic genes, their combination and interaction with tobacco smoke and alcohol consumption and risk of gastric cancer: A case-control study in an Italian population. BMC Cancer 2007, 7, 206. [Google Scholar] [CrossRef]
- Chen, S.; Laverdiere, I.; Tourancheau, A.; Jonker, D.; Couture, F.; Cecchin, E.; Villeneuve, L.; Harvey, M.; Court, M.H.; Innocenti, F.; et al. A novel UGT1 marker associated with better tolerance against irinotecan-induced severe neutropenia in metastatic colorectal cancer patients. Pharm. J. 2015, 15, 513–520. [Google Scholar] [CrossRef]
- Tetik Vardarli, A.; Harman, E.; Bozok Cetintas, V.; Kayikcioglu, M.; Vardarli, E.; Zengi, A.; Kucukaslan, A.S.; Eroglu, Z. Polymorphisms of lipid metabolism enzyme-coding genes in patients with diabetic dyslipidemia. Anatol. J. Cardiol. 2017, 17, 313–321. [Google Scholar] [CrossRef]
- Zakrzewski-Jakubiak, M.; de Denus, S.; Dube, M.P.; Belanger, F.; White, M.; Turgeon, J. Ten renin-angiotensin system-related gene polymorphisms in maximally treated Canadian Caucasian patients with heart failure. Br. J. Clin. Pharm. 2008, 65, 742–751. [Google Scholar] [CrossRef] [Green Version]
- Roszak, A.; Lutkowska, A.; Lianeri, M.; Sowinska, A.; Jagodzinski, P.P. Involvement of myeloperoxidase gene polymorphism 463G > A in development of cervical squamous cell carcinoma. Int. J. Biol. Markers 2016, 31, 440–445. [Google Scholar] [CrossRef]
- Mazzuca, F.; Borro, M.; Botticelli, A.; Aimati, L.; Gentile, G.; Capalbo, C.; Maddalena, C.; Mazzotti, E.; Simmaco, M.; Marchetti, P. Effect of MTHFR Polymorphisms on Gastrointestinal Cancer Risk in Italy. World J. Oncol. 2015, 6, 394–397. [Google Scholar] [CrossRef]
- Moreno, V.; Gemignani, F.; Landi, S.; Gioia-Patricola, L.; Chabrier, A.; Blanco, I.; Gonzalez, S.; Guino, E.; Capella, G.; Canzian, F. Polymorphisms in genes of nucleotide and base excision repair: Risk and prognosis of colorectal cancer. Clin. Cancer Res. 2006, 12, 2101–2108. [Google Scholar] [CrossRef] [Green Version]
- Micarelli, A.; Viziano, A.; Bruno, E.; Micarelli, E.; Alessandrini, M. Vestibular impairment in Multiple Chemical Sensitivity: Component analysis findings. J. Vestib. Res. 2016, 26, 459–468. [Google Scholar] [CrossRef]
- Micarelli, A.; Viziano, A.; Genovesi, G.; Bruno, E.; Ottaviani, F.; Alessandrini, M. Lack of contralateral suppression in transient-evoked otoacoustic emissions in multiple chemical sensitivity: A clinical correlation study. Noise Health 2016, 18, 143–149. [Google Scholar] [CrossRef]
- Viziano, A.; Micarelli, A.; Alessandrini, M. Noise sensitivity and hyperacusis in patients affected by multiple chemical sensitivity. Int. Arch. Occup. Environ. Health 2017, 90, 189–196. [Google Scholar] [CrossRef]
- Casarett, L.; Doull, J. Casarett and Doull’s Toxicology: The Basic Science of Poisons, 7th ed.; Klaassen, K.D., Ed.; McGraw-Hill—Medical Publishing Division: New York, NY, USA, 2008; p. 1309. [Google Scholar] [CrossRef]
- McFadden, S.A. Phenotypic variation in xenobiotic metabolism and adverse environmental response: Focus on sulfur-dependent detoxification pathways. Toxicology 1996, 111, 43–65. [Google Scholar] [CrossRef]
- Weber, W.W.; Hein, D.W. N-acetylation pharmacogenetics. Pharm. Rev. 1985, 37, 25–79. [Google Scholar]
- Bouayed, J.; Rammal, H.; Soulimani, R. Oxidative stress and anxiety: Relationship and cellular pathways. Oxid. Med. Cell. Longev. 2009, 2, 63–67. [Google Scholar] [CrossRef]
- Ng, F.; Berk, M.; Dean, O.; Bush, A.I. Oxidative stress in psychiatric disorders: Evidence base and therapeutic implications. Int. J. Neuropsychopharmacol. 2008, 11, 851–876. [Google Scholar] [CrossRef] [Green Version]
- Ross, P.M.; Whysner, J.; Covello, V.T.; Kuschner, M.; Rifkind, A.B.; Sedler, M.J.; Trichopoulos, D.; Williams, G.M. Olfaction and symptoms in the multiple chemical sensitivities syndrome. Prev. Med. 1999, 28, 467–480. [Google Scholar] [CrossRef]
- Lee, Y.L.; Pai, M.C.; Chen, J.H.; Guo, Y.L. Central neurological abnormalities and multiple chemical sensitivity caused by chronic toluene exposure. Occup. Med. (Lond) 2003, 53, 479–482. [Google Scholar] [CrossRef] [Green Version]
- Pall, M.L. NMDA sensitization and stimulation by peroxynitrite, nitric oxide, and organic solvents as the mechanism of chemical sensitivity in multiple chemical sensitivity. FASEB J. 2002, 16, 1407–1417. [Google Scholar] [CrossRef]
- Bell, I.R.; Miller, C.S.; Schwartz, G.E. An olfactory-limbic model of multiple chemical sensitivity syndrome: Possible relationships to kindling and affective spectrum disorders. Biol. Psychiatry 1992, 32, 218–242. [Google Scholar] [CrossRef]
- Cabungcal, J.H.; Preissmann, D.; Delseth, C.; Cuenod, M.; Do, K.Q.; Schenk, F. Transitory glutathione deficit during brain development induces cognitive impairment in juvenile and adult rats: Relevance to schizophrenia. Neurobiol. Dis. 2007, 26, 634–645. [Google Scholar] [CrossRef]
- Farooqui, T. A potential link among biogenic amines-based pesticides, learning and memory, and colony collapse disorder: A unique hypothesis. Neurochem. Int. 2013, 62, 122–136. [Google Scholar] [CrossRef]
- Gardiner, J.; Barton, D.; Overall, R.; Marc, J. Neurotrophic support and oxidative stress: Converging effects in the normal and diseased nervous system. Neuroscientist 2009, 15, 47–61. [Google Scholar] [CrossRef]
- Kako, H.; Fukumoto, S.; Kobayashi, Y.; Yokogoshi, H. Effects of direct exposure of green odour components on dopamine release from rat brain striatal slices and PC12 cells. Brain Res. Bull. 2008, 75, 706–712. [Google Scholar] [CrossRef]
- Angelucci, F.L.; Silva, V.V.; Dal Pizzol, C.; Spir, L.G.; Praes, C.E.; Maibach, H. Physiological effect of olfactory stimuli inhalation in humans: An overview. Int. J. Cosmet. Sci. 2014, 36, 117–123. [Google Scholar] [CrossRef]
- Hojo, S.; Sakabe, K.; Ishikawa, S.; Miyata, M.; Kumano, H. Evaluation of subjective symptoms of Japanese patients with multiple chemical sensitivity using QEESI(c). Environ. Health Prev. Med. 2009, 14, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Robert-Hazotte, A.; Faure, P.; Neiers, F.; Potin, C.; Artur, Y.; Coureaud, G.; Heydel, J.M. Nasal mucus glutathione transferase activity and impact on olfactory perception and neonatal behavior. Sci. Rep. 2019, 9, 3104. [Google Scholar] [CrossRef]
- Hayes, J.D.; Pulford, D.J. The glutathione S-transferase supergene family: Regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 445–600. [Google Scholar] [CrossRef]
- Saito, M.; Kumano, H.; Yoshiuchi, K.; Kokubo, N.; Ohashi, K.; Yamamoto, Y.; Shinohara, N.; Yanagisawa, Y.; Sakabe, K.; Miyata, M.; et al. Symptom profile of multiple chemical sensitivity in actual life. Psychosom. Med. 2005, 67, 318–325. [Google Scholar] [CrossRef] [Green Version]
- La Du, B.N.; Aviram, M.; Billecke, S.; Navab, M.; Primo-Parmo, S.; Sorenson, R.C.; Standiford, T.J. On the physiological role(s) of the paraoxonases. Chem. Biol. Interact. 1999, 119–120, 379–388. [Google Scholar] [CrossRef]
- Billecke, S.; Draganov, D.; Counsell, R.; Stetson, P.; Watson, C.; Hsu, C.; La Du, B.N. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab. Dispos. 2000, 28, 1335–1342. [Google Scholar]
- Jakubowski, H. Calcium-dependent human serum homocysteine thiolactone hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 2000, 275, 3957–3962. [Google Scholar] [CrossRef] [Green Version]
- Smolen, A.; Eckerson, H.W.; Gan, K.N.; Hailat, N.; La Du, B.N. Characteristics of the genetically determined allozymic forms of human serum paraoxonase/arylesterase. Drug Metab. Dispos. 1991, 19, 107–112. [Google Scholar] [PubMed]
- Di Mauro, R.; Cantarella, G.; Bernardini, R.; Di Rosa, M.; Barbagallo, I.; Distefano, A.; Longhitano, L.; Vicario, N.; Nicolosi, D.; Lazzarino, G.; et al. The Biochemical and Pharmacological Properties of Ozone: The Smell of Protection in Acute and Chronic Diseases. Int. J. Mol. Sci. 2019, 20, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, C.; Scordo, G.; Cesareo, E.; Raskovic, D.; Genovesi, G.; Korkina, L. Idiopathic environmental intolerances (IEI): From molecular epidemiology to molecular medicine. Indian J. Exp. Biol. 2010, 48, 625–635. [Google Scholar] [PubMed]
- Holla, L.I.; Stejskalova, A.; Znojil, V.; Vasku, A. Analysis of the inducible nitric oxide synthase gene polymorphisms in Czech patients with atopic diseases. Clin. Exp. Allergy 2006, 36, 1592–1601. [Google Scholar] [CrossRef]
- Konno, S.; Hizawa, N.; Yamaguchi, E.; Jinushi, E.; Nishimura, M. (CCTTT)n repeat polymorphism in the NOS2 gene promoter is associated with atopy. J. Allergy Clin. Immunol. 2001, 108, 810–814. [Google Scholar] [CrossRef]
- Thiebaud, N.; Veloso Da Silva, S.; Jakob, I.; Sicard, G.; Chevalier, J.; Menetrier, F.; Berdeaux, O.; Artur, Y.; Heydel, J.M.; Le Bon, A.M. Odorant metabolism catalyzed by olfactory mucosal enzymes influences peripheral olfactory responses in rats. PLoS ONE 2013, 8, e59547. [Google Scholar] [CrossRef] [Green Version]
- Asakawa, M.; Fukutani, Y.; Savangsuksa, A.; Noguchi, K.; Matsunami, H.; Yohda, M. Modification of the response of olfactory receptors to acetophenone by CYP1a2. Sci. Rep. 2017, 7, 10167. [Google Scholar] [CrossRef] [Green Version]
- Heydel, J.M.; Menetrier, F.; Belloir, C.; Canon, F.; Faure, P.; Lirussi, F.; Chavanne, E.; Saliou, J.M.; Artur, Y.; Canivenc-Lavier, M.C.; et al. Characterization of rat glutathione transferases in olfactory epithelium and mucus. PLoS ONE 2019, 14, e0220259. [Google Scholar] [CrossRef] [Green Version]
- Miksys, S.; Rao, Y.; Hoffmann, E.; Mash, D.C.; Tyndale, R.F. Regional and cellular expression of CYP2D6 in human brain: Higher levels in alcoholics. J. Neurochem. 2002, 82, 1376–1387. [Google Scholar] [CrossRef]
- Gaedigk, A.; Gotschall, R.R.; Forbes, N.S.; Simon, S.D.; Kearns, G.L.; Leeder, J.S. Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics 1999, 9, 669–682. [Google Scholar] [CrossRef]
- Yu, A.M.; Idle, J.R.; Krausz, K.W.; Kupfer, A.; Gonzalez, F.J. Contribution of individual cytochrome P450 isozymes to the O-demethylation of the psychotropic beta-carboline alkaloids harmaline and harmine. J. Pharm. Exp. Ther. 2003, 305, 315–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llerena, A.; Edman, G.; Cobaleda, J.; Benitez, J.; Schalling, D.; Bertilsson, L. Relationship between personality and debrisoquine hydroxylation capacity. Suggestion of an endogenous neuroactive substrate or product of the cytochrome P4502D6. Acta Psychiatr. Scand. 1993, 87, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Miksys, S.; Rao, Y.; Sellers, E.M.; Kwan, M.; Mendis, D.; Tyndale, R.F. Regional and cellular distribution of CYP2D subfamily members in rat brain. Xenobiotica 2000, 30, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Multiple chemical sensitivity: A 1999 consensus. Arch. Environ. Health 1999, 54, 147–149. [CrossRef] [PubMed]
- Lacour, M.; Zunder, T.; Schmidtke, K.; Vaith, P.; Scheidt, C. Multiple chemical sensitivity syndrome (MCS)—suggestions for an extension of the U.S. MCS-case definition. Int. J. Hyg. Environ. Health 2005, 208, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Tempere, S.; Hamtat, M.L.; Bougeant, J.C.; de Revel, G.; Sicard, G. Learning Odors: The Impact of Visual and Olfactory Mental Imagery Training on Odor Perception. J. Sens. Stud. 2014, 29, 435–449. [Google Scholar] [CrossRef]
- Ferlazzo, N.; Gorgone, G.; Caccamo, D.; Curro, M.; Condello, S.; Pisani, F.; Vernieri, F.; Rossini, P.M.; Ientile, R. The 894G > T (Glu298Asp) variant in the endothelial NOS gene and MTHFR polymorphisms influence homocysteine levels in patients with cognitive decline. Neuromol. Med. 2011, 13, 167–174. [Google Scholar] [CrossRef]
- Vecchio, M.; Curro, M.; Trimarchi, F.; Naccari, S.; Caccamo, D.; Ientile, R.; Barreca, D.; Di Mauro, D. The Oxidative Stress Response in Elite Water Polo Players: Effects of Genetic Background. Biomed. Res. Int. 2017, 2017, 7019694. [Google Scholar] [CrossRef]
- Di Mauro, D.; Curro, M.; Trimarchi, F.; Vecchio, M.; Rizzo, G.; Barreca, D.; Visalli, G.; Ientile, R.; Caccamo, D. Role of Genetic Background in Cardiovascular Risk Markers Changes in Water Polo Players. Int. J. Sports Med. 2018, 39, 390–396. [Google Scholar] [CrossRef]
- Massacesi, C.; Terrazzino, S.; Marcucci, F.; Rocchi, M.B.; Lippe, P.; Bisonni, R.; Lombardo, M.; Pilone, A.; Mattioli, R.; Leon, A. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer 2006, 106, 1007–1016. [Google Scholar] [CrossRef]
- Pandolfo, G.; Gugliandolo, A.; Gangemi, C.; Arrigo, R.; Curro, M.; La Ciura, G.; Muscatello, M.R.; Bruno, A.; Zoccali, R.; Caccamo, D. Association of the COMT synonymous polymorphism Leu136Leu and missense variant Val158Met with mood disorders. J. Affect. Disord. 2015, 177, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, E.; Katotomichelakis, M.; Gouveris, H.; Tripsianis, G.; Livaditis, M.; Danielides, V. Olfaction-associated quality of life in chronic rhinosinusitis: Adaptation and validation of an olfaction-specific questionnaire. Laryngoscope 2012, 122, 1450–1454. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, M.; Micarelli, A.; Chiaravalloti, A.; Candidi, M.; Bruno, E.; Di Pietro, B.; Oberg, J.; Schillaci, O.; Pagani, M. Cerebellar metabolic involvement and its correlations with clinical parameters in vestibular neuritis. J. Neurol. 2014, 261, 1976–1985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funke, S.; Risch, A.; Nieters, A.; Hoffmeister, M.; Stegmaier, C.; Seiler, C.M.; Brenner, H.; Chang-Claude, J. Genetic Polymorphisms in Genes Related to Oxidative Stress (GSTP1, GSTM1, GSTT1, CAT, MnSOD, MPO, eNOS) and Survival of Rectal Cancer Patients after Radiotherapy. J. Cancer Epidemiol. 2009, 2009, 302047. [Google Scholar] [CrossRef] [Green Version]
- Micarelli, A.; Liguori, C.; Viziano, A.; Izzi, F.; Placidi, F.; Alessandrini, M. Integrating postural and vestibular dimensions to depict impairment in moderate-to-severe obstructive sleep apnea syndrome patients. J. Sleep Res. 2017, 26, 487–494. [Google Scholar] [CrossRef] [Green Version]
- Micarelli, A.; Viziano, A.; Panella, M.; Micarelli, E.; Alessandrini, M. Power spectra prognostic aspects of impulsive eye movement traces in superior vestibular neuritis. Med. Biol. Eng. Comput. 2019, 57, 1617–1627. [Google Scholar] [CrossRef]
- Park, S.H.; Park, J.O. Simultaneous Optimization of Multiple Responses Using a Weighted Desirability Function. In Quality Improvement Through Statistical Methods; Abraham, B., Ed.; Birkhäuser Boston: Boston, MA, USA, 1998; pp. 299–311. [Google Scholar] [CrossRef]
- Sakurai, A.; Ihara, S.; Tagami, R.; Yamaguchi, J.; Sugita, A.; Kuwana, T.; Sawada, N.; Hori, S.; Taniguch, T.; Kinoshita, K. Parameters Influencing Brain Oxygen Measurement by Regional Oxygen Saturation in Postcardiac Arrest Patients with Targeted Temperature Management. Ther. Hypothermia Temp. Manag. 2019. [Google Scholar] [CrossRef] [Green Version]
- Halili, L.; Liu, R.H.; Weeks, A.; Deonandan, R.; Adamo, K.B. High maternal self-efficacy is associated with meeting Institute of Medicine gestational weight gain recommendations. PLoS ONE 2019, 14, e0226301. [Google Scholar] [CrossRef] [Green Version]

| QOD NS | 23.8 ± 4.56 |
| QOD PS | 4.86 ± 0.97 |
| LQrv (NS + PS) | 28.67 ± 4.22 |
| Age (years) | 47.23 ± 10.06 |
| Gender | 27 females; 19 males |
| Compounds exposure | 24 |
| Psychological trauma | 17 |
| Physical trauma | 14 |
| Previous surgery | 14 |
| Gene Polymorphism | Genotype/Alleles | Genotype and Allele Frequency in MCS Patients | Genotype and Allele Frequency in Caucasian Population | p |
|---|---|---|---|---|
| CYP2C9 C430T, *2 (R144C) A1075T, *3 (I359L) | CC | 63.0% | 80% §1 | 0.0273 |
| CT | 34.8% | 17.2% §1 | 0.0044 | |
| TT | 2.2% | 2.8% §1 | 0.9833 | |
| C(*1), T(*2) | 0.804, 0.196 | 0.886, 0.124 §1 | ||
| AA | 82.6% | 98.9% §1 | <0.00001 | |
| AT | 17.4% | 1.1% §1 | <0.00001 | |
| TT | 0% | 0% §1 | - | |
| A(*1), T(*3) | 0.913, 0.087 | 0.994, 0.056 §1 | ||
| *2/*3 | 10.9% | 2.2% §1 | 0.0097 | |
| CYP2C19 G681A, *2 -C806T, *17 | *1/*1 | 43.5% | 49.2% §2 | 0.252 |
| *1/*2 | 21.7% | 16.4% §2 | 0.279 | |
| *2/*2 | 2.2% | 2.8% §2 | 1 | |
| *1/*17 | 30.4% | 22.8% §2 | 0.179 | |
| *17/*17 | 2.2% | 2.8 §2 | 1 | |
| *1, *2, *17 | 0.695, 0.130, 0.175 | 0.688, 0.110, 0.142 §2 | ||
| CYP2D6 C2850T, *2 (R296C) -C1584G, *2A G1846A, *4 1707delT, *6 C100T, *10 G2988A, *41 | PM # (4*10/4*10) | 6.5% | 6.7% §2 | 1.0 |
| IM # (*1/*4/*10, *2*2A/*4*10 *10/*4*10) | 6.5% | 28.9% §2 | 0.006 | |
| EM # (1*/*1, *2/*2, *2A/*2A, *2*41/*2*2A *2*4*10/*2A, *4/*10, *1/*6) | 80.5% | 62.2% §2 | 0.154 | |
| UM # (*2/*2A*XN) | 6.5% | 2.2% §2 | 0.617 | |
| GSTP1 A313G | AA | 41.3% | 23.5% §4 | 0.0074 |
| AG | 50.0% | 62.5% §4 | 0.0943 | |
| GG | 8.7% | 14% §4 | 0.4888 | |
| A, G | 0.663, 0.337 | 0.70, 0.30 §4 | 0.4573 | |
| GSTM1 DEL | INS, DEL | 0.413, 0.587 | 0.527, 0.473, §5 | 0.4573 |
| GSTT1 DEL | INS, DEL | 0.761, 0.239 | 0.78, 0.22 §5 | 0.8263 |
| GSTM1/GSTT1 | INS/DEL | 15% | 15% §3 | 0.96 |
| GSTM1/GSTT1 | DEL/INS | 50.3% | 41% §3 | 0.25 |
| GSTM1/GSTT1 | DEL/DEL | 9.0% | 1.5% §3 | 0.85 |
| UGT1A1 (TA)7TAA, *28 | *1/*1 | 32.6% | 48.5% §6 | 0.0532 |
| *1/*28 | 52.2% | 39.0% §6 | 0.135 | |
| *28/*28 | 15.2% | 12.5% §6 | 0.631 | |
| *1, *28 | 0.587, 0.413 | 0.701, 0.299 §6 | ||
| Gene Polymorphism (Amino Acid Substitution) | Genotype/Alleles | Genotype and Allele Frequency in MCS Patients | Genotype and Allele Frequency in Caucasian Population | p |
| SOD2 C48T (A16V) | CC | 19.6% | 40.2% §7 | 0.0076 |
| CT | 47.8% | 45.2% §7 | 0.749 | |
| TT | 32.6% | 14.6% §7 | 0.0056 | |
| C, T | 0.435, 0.565 | 0.628, 0.372 §7 | ||
| CAT -C262T | CC | 56.5% | 80% §8 | 0.0008 |
| CT | 32.6% | 19% §8 | 0.0415 | |
| TT | 10.9% | 1% §8 | 0.0004 | |
| C, T | 0.728, 0.272 | 0.895, 0.105 §8 | ||
| PON1 A575G (Q192R) C108T (L55M) | AA | 50% | 52.1% §9 | 0.4485 |
| AG | 39.1% | 36.3% §9 | 0.7202 | |
| GG | 10.9% | 7.6% §9 | 0.4551 | |
| A, G | 0.696, 0.304 | 0.742, 0.258 §9 | ||
| CC | 28.3% | 19.9% §4 | 0.3868 | |
| CT | 47.8% | 54.4% §4 | 0.0002 | |
| TT | 23.9% | 26.7% §4 | 0.014 | |
| C, T | 0.522, 0.478 | 0.461, 0.539 §4 | ||
| AG/CT | 10.9% | / | ||
| NOS3 G894T (D298E) | GG | 45.7% | 36.9% §10 | 0.3093 |
| GT | 32.6% | 47.8% §10 | 0.0814 | |
| TT | 21.7% | 15.3% §10 | 0.3316 | |
| G, T | 0.619, 0.381 | 0.608, 0.392 §10 | ||
| MPO G463A | GG | 43.5% | 64.9 §11 | 0.057 |
| GA | 45.6% | 35% §11 | 0.1535 | |
| AA | 10.9% | 0.1% §11 | 0.4436 | |
| G, A | 0.337, 0.663 | 0.76, 0.24 §11 | ||
| MTHFR C677T (A222V) A1298C (E429A) | CC | 34.8% | 37.6% §12 | 0.7188 |
| CT | 43.5% | 48.5% §12 | 0.3766 | |
| TT | 23.9% | 13.9% §12 | 0.091 | |
| C, T | 0.543, 0.457 | 0.619, 0.381 §12 | ||
| AA | 41.3% | 51% §12 | 0.256 | |
| AC | 47.8% | 39.6% §12 | 0.323 | |
| CC | 10.9% | 9.4% §12 | 0.783 | |
| A, C | 0.772, 0.228 | 0.708, 0.292 §12 | ||
| CT/AC | 23.9% | 18.4%§4 | 0.505 | |
| OGG1 C315G (S326C) | CC | 78.3% | 65.0% §13 | 0.094 |
| CG | 19.6% | 32.2% §13 | 0.0893 | |
| GG | 2.2% | 2.8% §13 | 1 | |
| C, G | 0.804, 0.196 | 0.811, 0.189 §13 |
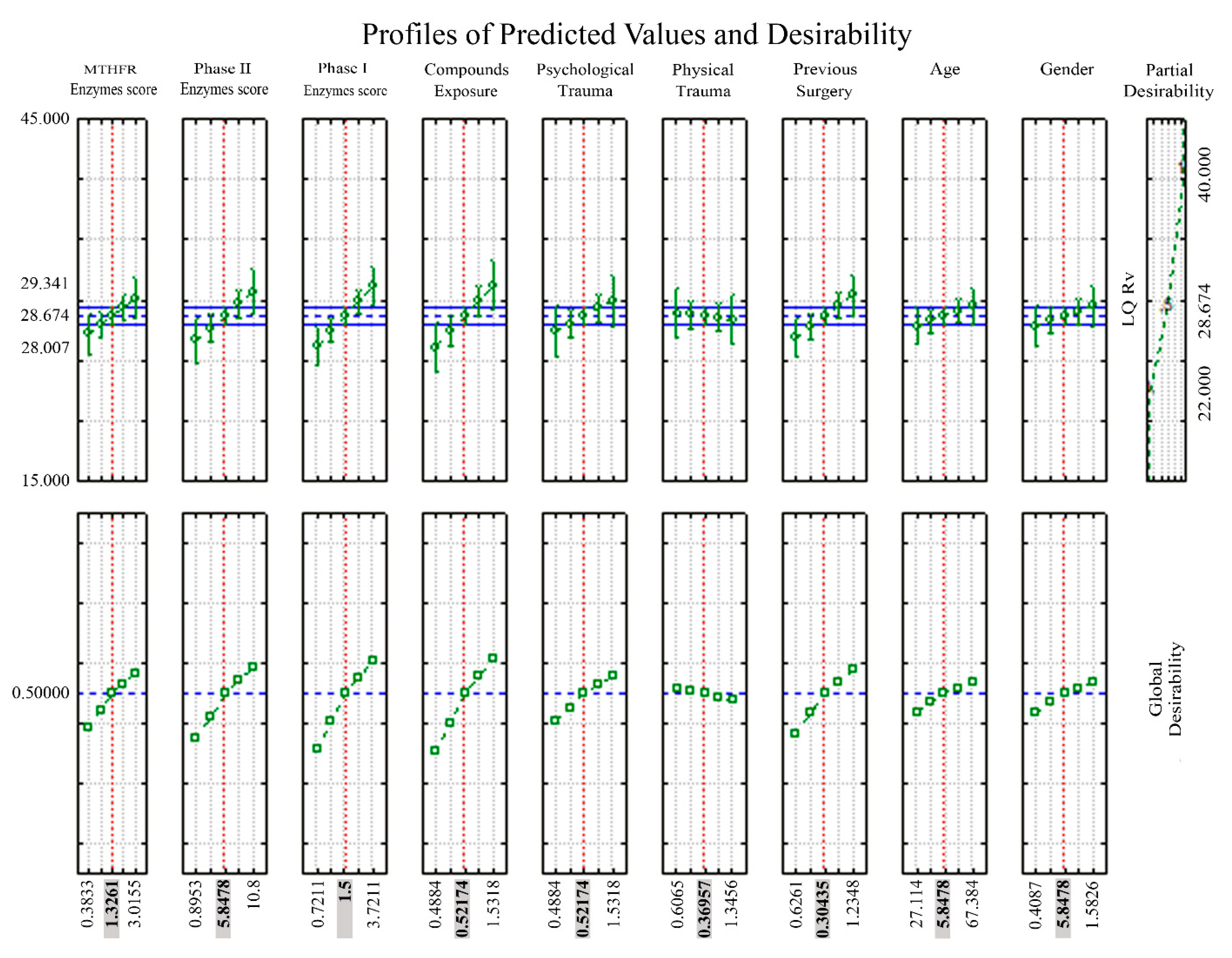
| Partial Regression Coefficient | Std.Err | t | p-Value | Cnf.Lmt −95.00% | Cnf.Lmt +95.00% | Partial Correlation Coefficient (ß) | Std.Err. ß | Cnf.Lmt −95.00% | Cnf.Lmt +95.00% | |
|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | 18.63568 | 1.953092 | 9.541631 | 0.000000 | 14.67463 | 22.59673 | ||||
| Phase I enzymes score | 1.09827 | 0.316817 | 3.466581 | 0.001382 | 0.45574 | 1.74081 | 0.288895 | 0.083337 | 0.119879 | 0.457910 |
| Compounds Exposure | 2.53639 | 0.976863 | 2.596462 | 0.013552 | 0.55522 | 4.51756 | 0.303415 | 0.116857 | 0.066418 | 0.540412 |
| Previous Surgery | 1.89423 | 0.812950 | 2.330064 | 0.025525 | 0.24549 | 3.54296 | 0.208725 | 0.089579 | 0.027050 | 0.390400 |
| Phase II enzymes score | 0.40192 | 0.174334 | 2.305473 | 0.027012 | 0.04836 | 0.75549 | 0.235737 | 0.102251 | 0.028362 | 0.443113 |
| MTHFR enzymes score | 0.87996 | 0.454025 | 1.938130 | 0.060481 | −0.04085 | 1.80076 | 0.176058 | 0.090839 | −0.008172 | 0.360289 |
| Psychological Trauma | 1.25605 | 0.999760 | 1.256353 | 0.217080 | −0.77156 | 3.28366 | 0.150255 | 0.119596 | −0.092297 | 0.392807 |
| Age | 0.04117 | 0.034592 | 1.190050 | 0.241816 | −0.02899 | 0.11132 | 0.098115 | 0.082446 | −0.069093 | 0.265323 |
| Gender | 0.81441 | 0.777464 | 1.047519 | 0.301843 | −0.76236 | 2.39118 | 0.096030 | 0.091673 | −0.089893 | 0.281952 |
| Physical Trauma | −0.28221 | 0.952656 | −0.296230 | 0.768757 | −2.21428 | 1.64987 | −0.032621 | 0.110119 | −0.255953 | 0.190712 |
| Partial Desirability Values | |||||
|---|---|---|---|---|---|
| Prognostic Factor | Factor Level | Predicted LQrv | Desirability Value | −95% CI LQrv | +95% CI LQrv |
| MTHFR enzymes score | −0.363323 | 27.18730 | 0.388626 | 25.49463 | 28.87997 |
| 0.4813819 | 27.93061 | 0.444314 | 26.90583 | 28.95539 | |
| 1.326087 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 2.170792 | 29.41722 | 0.532814 | 28.39244 | 30.44200 | |
| 3.015497 | 30.16052 | 0.565628 | 28.46785 | 31.85319 | |
| Phase II enzymes score | 0.8952978 | 26.68338 | 0.350873 | 24.80952 | 28.55724 |
| 3.371562 | 27.67865 | 0.425437 | 26.57786 | 28.77943 | |
| 5.847826 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 8.324090 | 29.66918 | 0.543937 | 28.56839 | 30.76996 | |
| 10.80035 | 30.66444 | 0.587874 | 28.79059 | 32.53830 | |
| Phase I enzymes score | −0.721111 | 26.23453 | 0.317246 | 24.65912 | 27.80994 |
| 0.3894446 | 27.45422 | 0.408623 | 26.47730 | 28.43114 | |
| 1.500000 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 2.610555 | 29.89360 | 0.553845 | 28.91669 | 30.87052 | |
| 3.721111 | 31.11329 | 0.607689 | 29.53789 | 32.68870 | |
| Compounds exposures | −0.488355 | 26.11192 | 0.308060 | 24.00246 | 28.22139 |
| 0.0166921 | 27.39292 | 0.404031 | 26.19027 | 28.59556 | |
| 0.5217391 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 1.026786 | 29.95491 | 0.556551 | 28.75226 | 31.15755 | |
| 1.531833 | 31.23590 | 0.613102 | 29.12643 | 33.34537 | |
| Psychological trauma | −0.488355 | 27.40518 | 0.404949 | 25.25116 | 29.55920 |
| 0.0166921 | 28.03955 | 0.452475 | 26.81732 | 29.26178 | |
| 0.5217391 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 1.026786 | 29.30828 | 0.528005 | 28.08605 | 30.53051 | |
| 1.531833 | 29.94264 | 0.556010 | 27.78862 | 32.09666 | |
| Physical Trauma | −0.606476 | 28.94936 | 0.512160 | 26.94901 | 30.94970 |
| −0.118456 | 28.81164 | 0.506080 | 27.65654 | 29.96673 | |
| 0.3695652 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 0.8575860 | 28.53619 | 0.489683 | 27.38110 | 29.69128 | |
| 1.345607 | 28.39847 | 0.479365 | 26.39813 | 30.39881 | |
| Previous Surgery | −0.626082 | 26.91147 | 0.367961 | 25.23861 | 28.58433 |
| −0.160867 | 27.79269 | 0.433981 | 26.77608 | 28.80931 | |
| 0.3043478 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 0.7695630 | 29.55514 | 0.538903 | 28.53852 | 30.57175 | |
| 1.234778 | 30.43636 | 0.577805 | 28.76350 | 32.10922 | |
| Age | 27.11425 | 27.84545 | 0.437933 | 26.28385 | 29.40705 |
| 37.17669 | 28.25968 | 0.468967 | 27.28832 | 29.23104 | |
| 47.23913 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 57.30157 | 29.08815 | 0.518287 | 28.11679 | 30.05950 | |
| 67.36401 | 29.50238 | 0.536574 | 27.94078 | 31.06398 | |
| Gender | −0.408686 | 27.86305 | 0.439252 | 26.15725 | 29.56886 |
| 0.0891352 | 28.26848 | 0.469627 | 27.23827 | 29.29869 | |
| 0.5869565 | 28.67391 | 0.500001 | 28.00669 | 29.34114 | |
| 1.084778 | 29.07934 | 0.517899 | 28.04913 | 30.10955 | |
| 1.582599 | 29.48477 | 0.535797 | 27.77897 | 31.19058 | |
| Enzyme Name | Gene Symbol | Catalyzed Reaction | Description of Enzyme Activity | Gene Polymorphisms Affecting Enzyme Function * |
|---|---|---|---|---|
| Catalase | CAT | 2 H2O2 ⇄ 2 H2O + O2 | Decomposition of hydrogen peroxide to water and oxygen, protecting the cells from oxidative damage by ROS. Catalase reactions are a key part of body’s enzymatic antioxidant defense mechanisms. | CAT-262C > T |
| Cytochrome P450 monoxygenase isozymes 2C9, 2C19, 2D6 | CYP2C9 CYP2C19 CYP2D6 | RH + O2 + NADPH + H+ → ROH + H2O + NADP+ | Insertion of one atom of oxygen (monoxygenation) into the aliphatic position of an organic substrate (RH), while the other oxygen atom is reduced to water. This biotransformation reaction is often referred to as “phase I detoxification” in xenobiotic metabolism. | CYP2C9 *2, *3 CYP2C19 *2, *17 CYP2D6 *4, *6, *10, *41 |
| Glutathione-S-transferase isozymes P1, M1, T1 | GSTP1 GSTM1 GSTT1 | GSH + X → GS-X + H+ | Conjugation of reduced glutathione (GSH) with toxic agents, carcinogens, drugs, leading to the formation of a detoxified complex more polar and more readily excreted from human body. This reaction is often referred to as “phase II detoxification” in xenobiotic metabolism. | GSTP1 I105V, A114V, GSTM1 null GSTT1 null |
| Methylenetetrahydrofolate reductase | MTHFR | 5,10-MTHF + NADPH → 5-MTHF+ NADP+ | Reduction of 5,10-methylenetetrahydrofolate (5,10-MTHF) to 5-methyltetrahydrofolate (5-MTHF), acting as methyl donor for homocysteine (Hcy) remethylation to methionine. Enzyme activity diminishment leads to Hcy accumulation that induces oxidative stress and endothelial dysfunction. | MTHFR C677T (A222V) MTHFR A1298C (E429V) |
| Myeloperoxidase | MPO | H2O2 + X− ⇄ H2O + HOX | MPO-mediated reaction of hydrogen peroxide with halide anions (X-),chloride, bromide, fluoride, iodide) generates hypohalous acids, that mediate the anti-microbial activity of neutrophil granulocytes, key cells of immune system. | MPO-G463A |
| Nitric oxide synthase type III | NOS3 | 2 L-Arginine + 3 NADPH + 3 H+ + 4 O2 ⇄ 2 Citrulline + 2 NO + 4 H2O + 3 NADP+ | Production of nitric oxide (NO) from L-arginine in the presence of NADPH, as cofactor, and oxygen. NO is a potent mediator of vasodilation in blood vessels. | NOS3 G894T (D298E) |
| 8-Oxoguanine glycosylase | OGG1 | 8-oxoG excision → nucleotide gap in DNA sequence | Excision of 8-oxoguanine (8-oxoG), an oxidized deoxyribonucleotide, with mutagenic effects, resulting from DNA exposure to ROS. OGG1-mediated cleavage of glycosidic bond causes a strand break in the DNA backbone. | OGG1 C315G (S326C) |
| Paraoxonase 1 | PON1 | OP + H2O → DEP + Phenol-X | Hydrolysis of pesticides organophosphates (OP) to diethylphosphate (DEP) and phenol compounds (Phenol-X: PNP, IMHP, TCP). PON1, as a HDL component, also protects against atherosclerosis by preventing the oxidation of plasma lipoproteins and the accumulation of oxidized LDLs and HDLs. | PON1 C108T (L55M), PON1 A575G (Q192R) |
| Superoxide dismutase 2 | SOD2 | 2O2− + 2H+ ⇄ O2 + H2O2 | Dismutation of superoxide radicals (O2−) to molecular oxygen (O2) and hydrogen peroxide (H2O2). SOD2 is a manganese-dependent enzyme located in mitochondria, providing to human cells a greatly effective defense against ROS. | SOD2 C28T (A16V) |
| UDP-glucuronosyl transferase isozyme 1A1 | UGT1A1 | UDP-GA + X → UDP + GA-X | Transfer of the glucuronic acid component of UDP-glucuronic acid (UDP-GA) to a small hydrophobic molecule (X) in microsomal compartment. Glucuronidation reaction takes place in phase II detoxification of xenobiotic metabolism. | UGT1A1 (TA)7TAA, * 28 |
| Odorants | Chemical Names | Odorants | Chemical Names |
|---|---|---|---|
| Almond | Benzaldehyde | Lavender | Essential oil |
| Anise | Anethol | Lemon | Citral |
| Apple | Ethyl 2-methylbutyrate | Mint | Essential oil |
| Banana | Isoamyl acetate | Mushroom | 1-Octen-3-one |
| Bell pepper | 2-Isobutyl-3-methoxypyrazine | Musk | Omega-pentadecalactone |
| Butter | Diacetyl | Onion | Ethanethiol |
| Cabbage | Methionol | Orange blossom | Methyl anthranilate |
| Camembert | S-methylthiobutyrate | Peach | γ-Undecalactone |
| Caramel | Furaneol | Pear | Hexyl acetate |
| Cinnamon | Cinnamaldehyde | Pineapple | Allyl hexanoate |
| Cloves | Eugenol | Plastic | Styrene |
| Coconut | Whiskey-lactone | Rose | Phenylethanol |
| Coffee | Extract | Rubber | Benzothiazole |
| Coriander | Linalool | Sea | Calone |
| Cork taint | 2,4,6-Trichloroanisole | Smokey | 4-Ethylgaïacol |
| Crab stick | Dimethylsulfide | Sweat | 3-Sulfanylhexyle acetate |
| Earthy | (±)-Geosmin | Thyme | Thymol |
| Eucalyptus | 1,8-Cineol | Toasted bread | 2-Acetylthiazole |
| Feet | Isovaleric acid | Vanilla | Aroma |
| Fish | 2-Butylamine | Vinegar | White vinegar |
| Grass | Cis-3-hexenol | Violet | β-Ionone |
| Horse | 4-Ethylphenol | Washing powder | Aldehyde |
| Kiwi | Ethyl butyrate | Woody | Cedryl acetate |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Micarelli, A.; Cormano, A.; Caccamo, D.; Alessandrini, M. Olfactory-Related Quality of Life in Multiple Chemical Sensitivity: A Genetic-Acquired Factors Model. Int. J. Mol. Sci. 2020, 21, 156. https://doi.org/10.3390/ijms21010156
Micarelli A, Cormano A, Caccamo D, Alessandrini M. Olfactory-Related Quality of Life in Multiple Chemical Sensitivity: A Genetic-Acquired Factors Model. International Journal of Molecular Sciences. 2020; 21(1):156. https://doi.org/10.3390/ijms21010156
Chicago/Turabian StyleMicarelli, Alessandro, Andrea Cormano, Daniela Caccamo, and Marco Alessandrini. 2020. "Olfactory-Related Quality of Life in Multiple Chemical Sensitivity: A Genetic-Acquired Factors Model" International Journal of Molecular Sciences 21, no. 1: 156. https://doi.org/10.3390/ijms21010156






