Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress
Abstract
:1. Introduction
2. Results
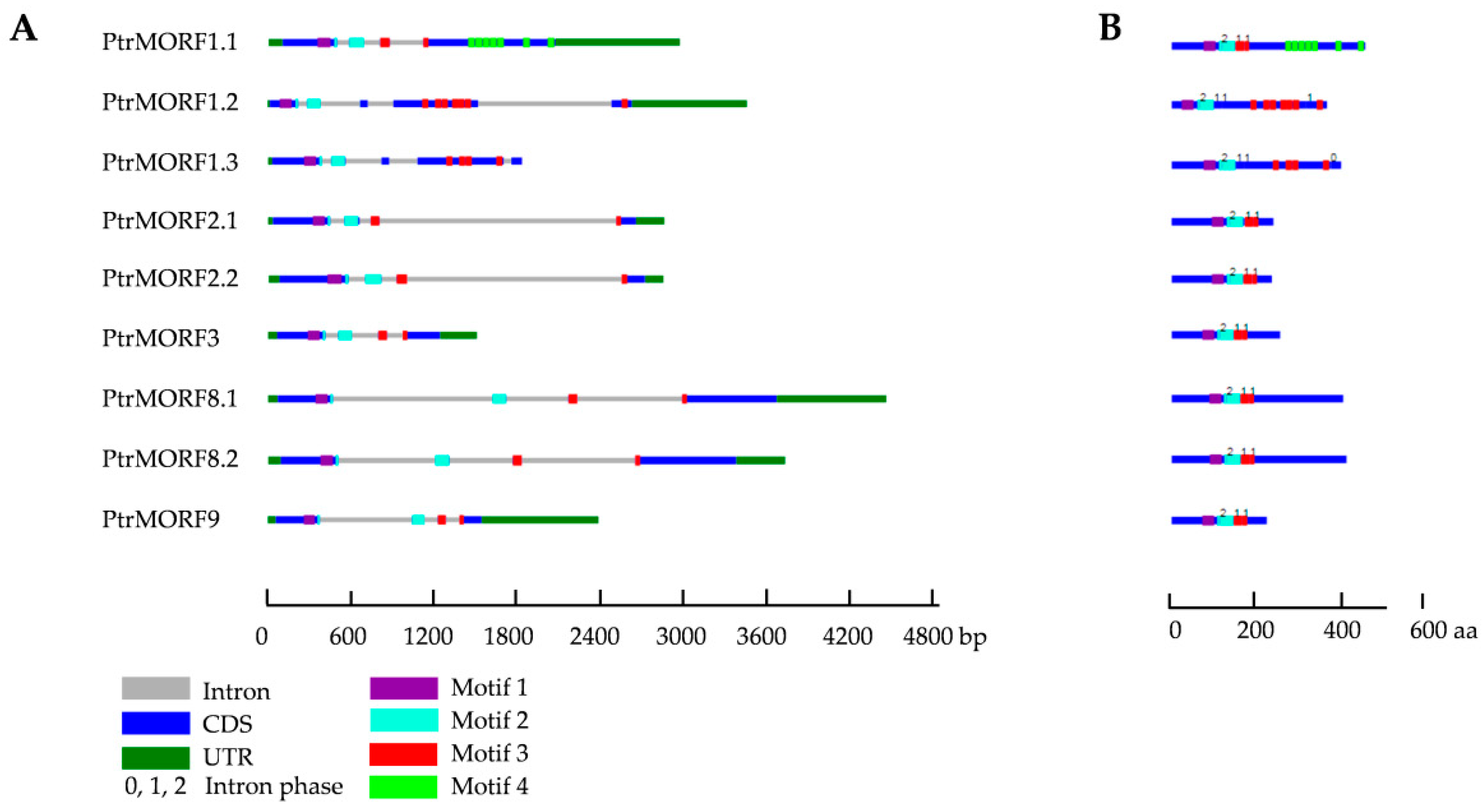
2.1. Identification and Sequence Analysis of the PtrMORF Gene Family
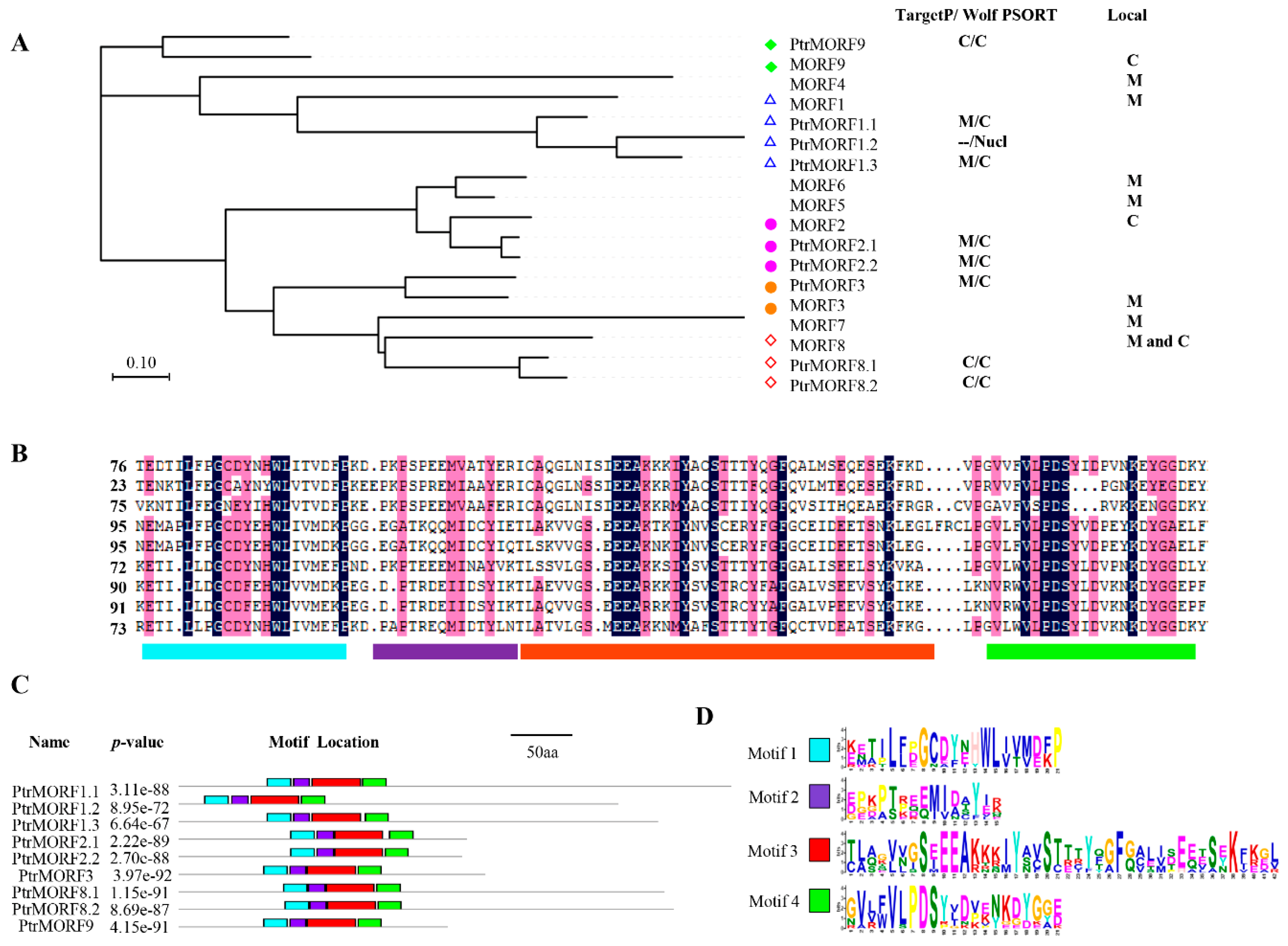
2.2. Phylogenetic Comparison of the MORF from Different Species
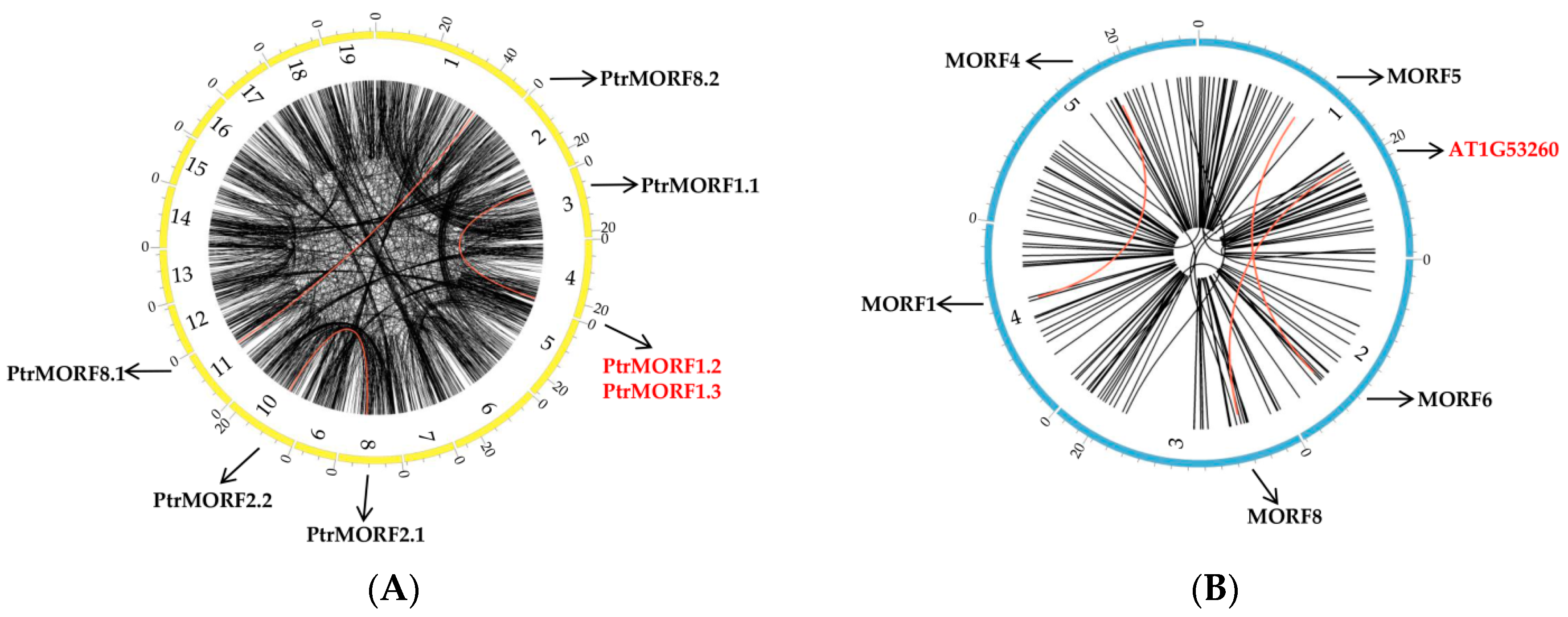
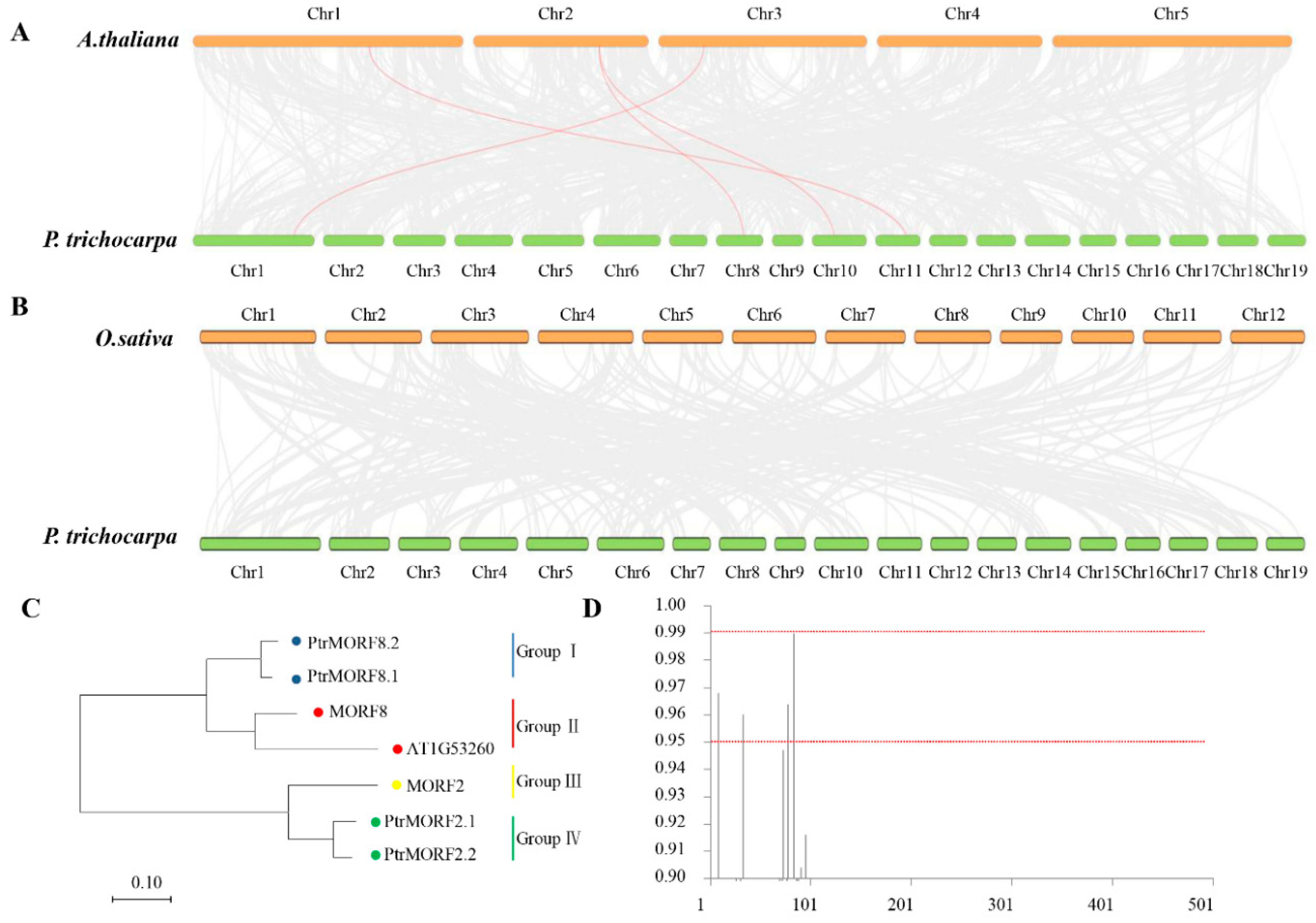
2.3. Chromosomal Distribution, Synteny, and Evolution of PtrMORF Genes
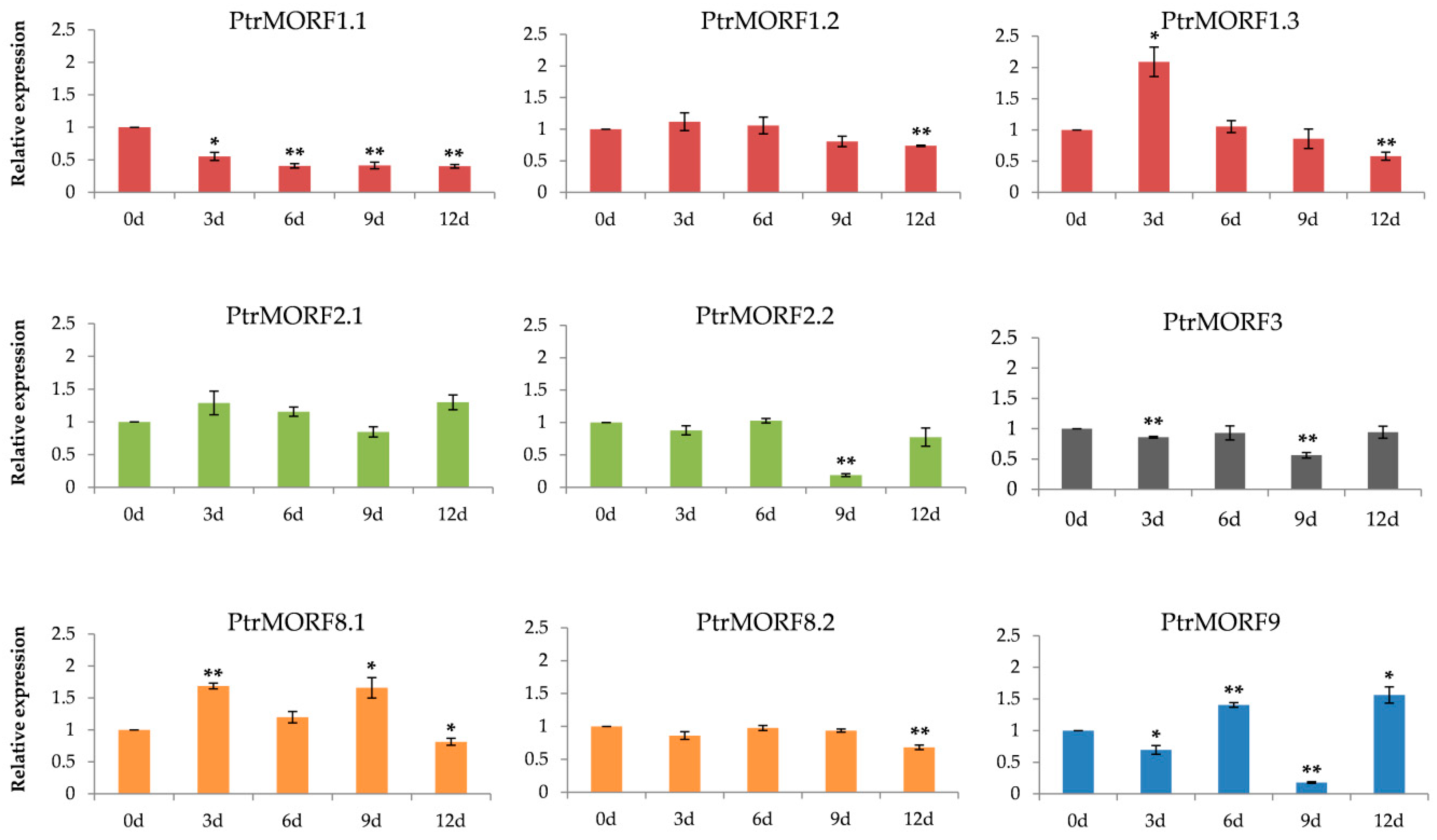
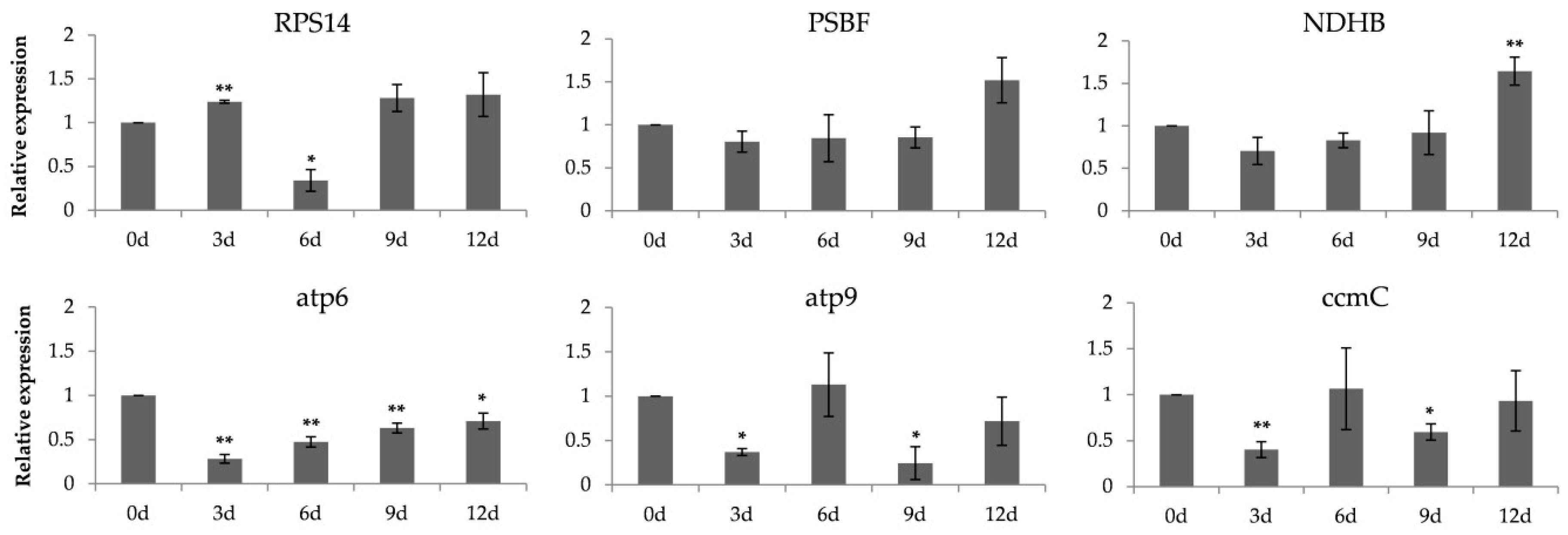
2.4. Expression of PtrMORF Genes and Six Genes from Chloroplasts and Mitochondria under Drought Stress
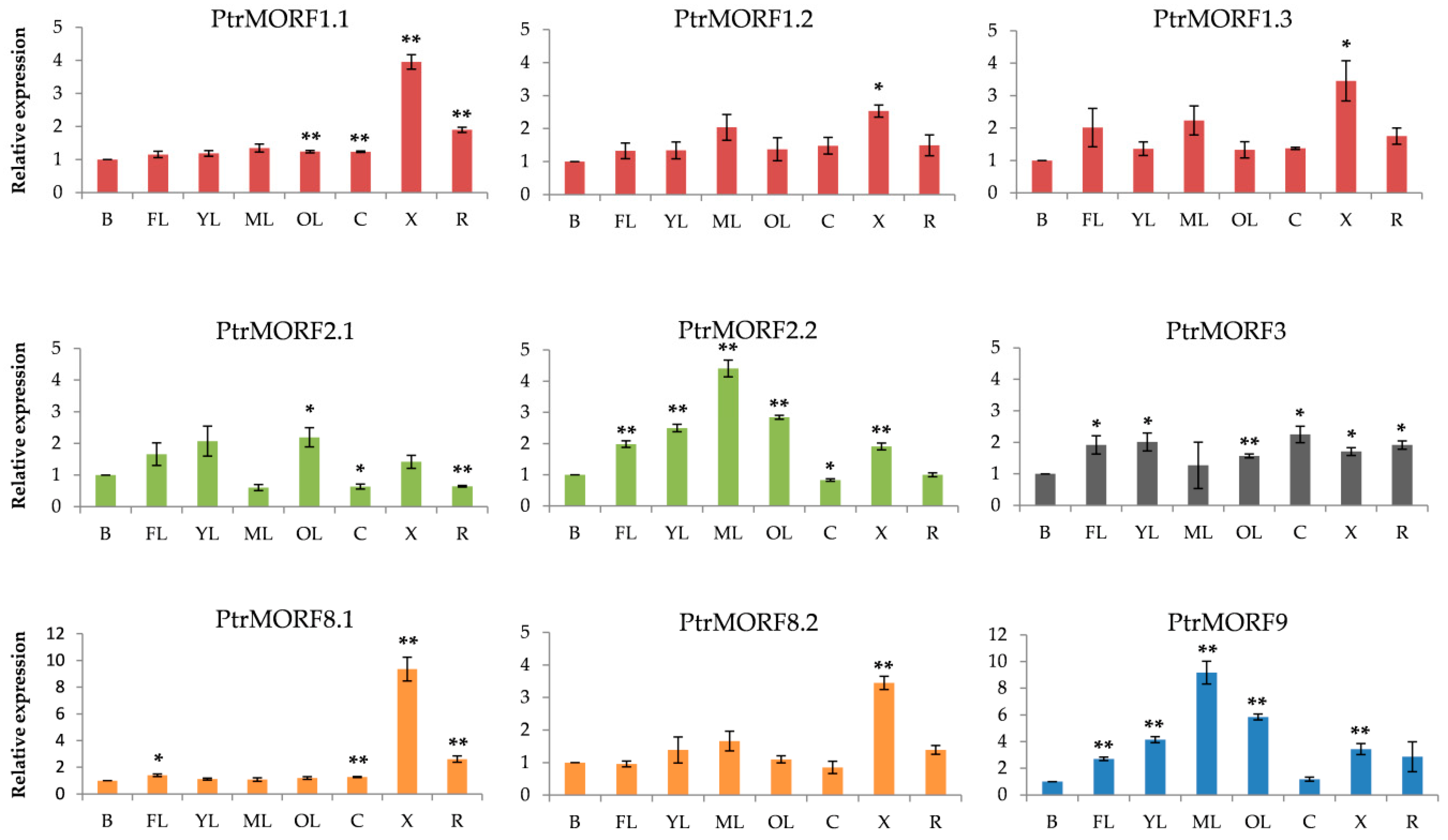
2.5. Expression Profiles of Populus MORF Gene
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Genome-Wide Identification and Sequence Analysis of MORF Genes in P. trichocarpa
4.3. Phylogenetic Analysis
4.4. Chromosome Location and Gene Structure Analysis
4.5. Colinearity of PtrMORF Gene
4.6. RT-PCR and qRT-PCR Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.P.; Teichmann, T. Poplar Genomics is Getting Popular: The Impact of the Poplar Genome Project on Tree Research. Plant Biol. 2004, 6, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, H.D.; Ceulemans, R.; Davis, J.; Stettler, R. Emerging Model Systems in Plant Biology: Poplar (Populus) as A Model Forest Tree. J. Plant Growth Regul. 2000, 19, 306–313. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, C.; Meng, S.; Li, M. Transcriptomic insight into nitrogen uptake and metabolism of Populus simonii in response to drought and low nitrogen stresses. Tree Physiol. 2018, 38, 1672–1684. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Bentolila, S.; Oh, J.; Hanson, M.R.; Bukowski, R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013, 9, e1003584. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yamaguchi, K.; Tsuji-Tsukinoki, S.; Knie, N.; Knoop, V. Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 2014, 20, 1499–1506. [Google Scholar] [CrossRef]
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant. Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Chateignerboutin, A.L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Freyer, R.; Kiefermeyer, M.C.; Kössel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA 1997, 94, 6285–6290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takenaka, M.; Neuwirt, J.; Brennicke, A. Complex cis-elements determine an RNA editing site in pea mitochondria. Nucleic Acids Res. 2004, 32, 4137–4144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Zhang, Q.; Yin, P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018, 1–8. [Google Scholar] [CrossRef]
- Bayer-Csaszar, E.; Haag, S.; Jorg, A.; Glass, F.; Hartel, B.; Obata, T.; Meyer, E.H.; Brennicke, A.; Takenaka, M. The conserved domain in MORF proteins has distinct affinities to the PPR and E elements in PPR RNA editing factors. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 813–828. [Google Scholar] [CrossRef]
- Shi, X.; Castandet, B.; Germain, A. ORRM5, an RNA recognition motif-containing protein, has a unique effect on mitochondrial RNA editing. J. Exp. Bot. 2017, 68, 2833–2847. [Google Scholar] [CrossRef] [Green Version]
- Hackett, J.B.; Shi, X.; Kobylarz, A.T.; Lucas, M.K.; Wessendorf, R.L.; Hines, K.M.; Bentolila, S.; Hanson, M.R.; Lu, Y. An Organelle RNA Recognition Motif Protein is Required for Photosynthetic Subunit psbF Transcript Editing. Plant. Physiol. 2017, 173, 2278–2293. [Google Scholar] [CrossRef]
- Shi, X.; Bentolila, S.; Hanson, M.R. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal. Behav. 2016, 11, 294–309. [Google Scholar] [CrossRef]
- Shi, X.; Hanson, M.R.; Bentolila, S. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 2015, 43, 3814–3825. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Tang, W.; Hedtke, B.; Zhong, L.; Liu, L.; Peng, L.; Lu, C.; Grimm, B.; Lin, R. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA 2014, 111, 2023–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Shi, X.; Friso, G.; Van Wijk, K.; Bentolila, S.; Hanson, M.R. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015, 11, e1005028. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Zhao, J.Y.; Lu, P.P.; Chen, M.; Guo, C.H.; Xu, Z.S.; Ma, Y.Z. The E-Subgroup Pentatricopeptide Repeat Protein Family in Arabidopsis thaliana and Confirmation of the Responsiveness PPR96 to Abiotic Stresses. Front. Plant Sci. 2016, 7, 1825. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kuhn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant. J. 2012, 71, 836–849. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Zhang, Q.; Guan, Z.; Wang, Q.; Li, L.; Ruan, F.; Lin, R.; Zou, T.; Yin, P. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants 2017, 3, 17037. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Hartel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Cai, M.; Zhang, J.; Li, Y.; Zhang, R.; Song, W.; Zhang, K.; Xiao, H.; Yue, B.; Zheng, Y.; et al. Functional divergencse and origin of the DAG-like gene family in plants. Sci. Rep. 2017, 7, 5688. [Google Scholar] [CrossRef]
- Zehrmann, A.; Hartel, B.; Glass, F.; Bayer-Csaszar, E.; Obata, T.; Meyer, E.; Brennicke, A.; Takenaka, M. Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 6445–6456. [Google Scholar] [CrossRef]
- Du, L.; Zhang, J.; Qu, S.; Zhao, Y.; Su, B.; Lv, X.; Li, R.; Wan, Y.; Xiao, J. The Pentratricopeptide Repeat Protein Pigment-Defective Mutant2 is Involved in the Regulation of Chloroplast Development and Chloroplast Gene Expression in Arabidopsis. Plant Cell Physiol. 2017, 58, 747–759. [Google Scholar] [CrossRef]
- Xiao, H.; Xu, Y.; Ni, C.; Zhang, Q.; Zhong, F.; Huang, J.; Liu, W.; Peng, L.; Zhu, Y.; Hu, J. A rice dual-localized pentatricopeptide repeat protein is involved in organellar RNA editing together with OsMORFs. J. Exp. Bot. 2018, 69, 2923–2936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, X.; Wang, Y.; Wu, J.; Gu, X.; Lu, T. The RNA Editing Factor WSP1 Is Essential for A Chloroplast Development in Rice. Mol. Plant 2017, 10, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Heller, W.P.; Sun, T.; Babina, A.M.; Friso, G.; Wijk, K.J.V.; Hanson, M.R. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA 2012, 109, 8372–8373. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Mattson, N.; Yang, L.; Jin, Q. Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: Evolution and differential expression during development and stress. BMC Genom. 2017, 18, 926. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-L.; Liu, Y.-J.; Wang, C.-L.; Zeng, Q.-Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Yuan, Y.; Gao, M.; Xing, L.; Li, C.; Li, M.; Ma, F. Genome-wide Identification, Classification, Molecular Evolution and Expression Analysis of Malate Dehydrogenases in Apple. Int. J. Mol. Sci. 2018, 19, 3312. [Google Scholar] [CrossRef]
- Xiong, J.; Tao, T.; Luo, Z.; Yan, S.; Liu, Y.; Yu, X.; Liu, G.; Xia, H.; Luo, L. RNA Editing Responses to Oxidative Stress between a Wild Abortive Type Male-Sterile Line and Its Maintainer Line. Front. Plant Sci. 2017, 8, 2023. [Google Scholar] [CrossRef]
- Rodrigues, N.F.; Fonseca, G.C.; Kulcheski, F.R.; Margis, R. Salt stress affects mRNA editing in soybean chloroplasts. Genet. Mol. Biol. 2017, 40, 200–208. [Google Scholar] [CrossRef] [Green Version]
- Yamburenko, M.V.; Zubo, Y.O.; Vanková, R.; Kusnetsov, V.V.; Kulaeva, O.N.; Börner, T. Abscisic acid represses the transcription of chloroplast genes. J. Exp. Bot. 2013, 64, 4491–4502. [Google Scholar] [CrossRef]
- Kurihara-Yonemoto, S.; Handa, H. Low temperature affects the processing pattern and RNA editing status of the mitochondrial cox2 transcripts in wheat. Curr. Genet. 2001, 40, 203–208. [Google Scholar] [CrossRef]
- Nakajima, Y.; Mulligan, R.M. Heat stress results in incomplete C-to-U editing of maize chloroplast mRNAs and correlates with changes in chloroplast transcription rate. Curr. Genet. 2001, 40, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Tan, Z.; Wu, F.; Sheng, P.; Heng, Y.; Wang, X.; Ren, Y.; Wang, J.; Guo, X.; Zhang, X.; et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 2014, 7, 1329–1349. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Sparvoli, S.; Edmunds, C.; Garosi, P.; Findlay, K.; Martin, C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996, 15, 4194–4207. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, C.; Begot, L.; Carol, P.; Perez, P.; Bligny, M.; Pesey, H.; Gallois, J.L.; Lerbs-Mache, S.; Mache, R. The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 2003, 51, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Ciuzan, O.; Hancock, J.; Pamfil, D.; Wilson, I.; Ladomery, M. The evolutionarily conserved multifunctional glycine-rich RNA-binding proteins play key roles in development and stress adaptation. Physiol. Plant 2015, 153, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Condit, C.M.; McLean, B.G.; Meagher, R.B. Characterization of the expression of the petunia glycine-rich protein-1 gene product. Plant Physiol. 1990, 93, 596–602. [Google Scholar] [CrossRef]
- Spicer, R. Symplasmic networks in secondary vascular tissues: Parenchyma distribution and activity supporting long-distance transport. J. Exp. Bot. 2014, 65, 1829–1848. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.; Xia, X.; Yin, W. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 2016, 14, 849–860. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; Von, H.G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adamscollier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Sandberg, G. The Populus Genome Integrative Explorer (PopGenIE): A new resource for exploring the Populus genome. New Phytol. 2009, 182, 1013–1025. [Google Scholar] [CrossRef]
- Liu, R.H.; Meng, J.L. MapDraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 2003, 25, 317–321. [Google Scholar]
- Wang, Y.; Xu, L.; Thilmony, R.; You, F.M.; Gu, Y.Q.; Colemanderr, D. PIECE 2.0: An update for the plant gene structure comparison and evolution database. Nucleic Acids Res. 2017, 45, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Model | P a | LnL b | Estimates of Parameters | ΔLRT c | df | p | Positively Selected Sites |
|---|---|---|---|---|---|---|---|
| M0 (one ratio model) | 1 | −1366.204 | ω = 0.055 | - | - | - | None |
| Branch-specific model (Model 2: two ratios) | |||||||
| Estimate ω for I | 2 | −1366.136 | ωI1 = 0.067, ω0 = 0.051 | Model 2 vs. M0: 0.136 | 1 | 0.712 | - |
| Estimate ω for II | 2 | −1357.726 | ωII1 = 0.177, ω0 = 0.028 | Model 2 vs. M0: 16.956 | 1 | 0.000 | - |
| Estimate ω for III | 2 | −1361.616 | ωIII1 = 0.018, ω0 = 0.079 | Model 2 vs. M0: 9.176 | 1 | 0.002 | - |
| Estimate ω for IV | 2 | −1363.519 | ωIV1 = 0.010, ω0 = 0.083 | Model 2 vs. M0: 5.370 | 1 | 0.002 | - |
| Model | P a | LnL b | Estimates of Parameters | ΔLRT c | df | p | Positively Selected Sites |
|---|---|---|---|---|---|---|---|
| M0 (one ratio model) | 1 | −1366.204 | ω = 0.0546 | - | - | - | None |
| Site-specific models | |||||||
| M1 (K = 2) | 1 | −1363.770 | p0 = 0.926, (p1 = 0.074) | - | - | - | Not allowed |
| M2 (K = 3) | 4 | −1363.770 | p0 = 0.926, p1 = 0.008, (p2 = 0.066), ω2 = 1.000 | M2 vs. M1: 0.000 | 2 | 1.000 | None |
| M3 (K = 8) | 5 | −1346.619 | p0 = 0.485, p1 = 0.001, p2 = 0.192, p3 = 0.174, p4 = 0.029, p5 = 0.040, p6 = 0.074, p7 = 0.007; ω0 = 0.006, ω1 = 0.062, ω2 = 0.062, ω3 = 0.062, ω4 = 0.266, ω5 = 0.266, ω6 = 0.266, ω7 = 0.266 | M3 vs. M0: 39.170 | 14 | 0.000 | None |
| M7 | 2 | −1347.154 | p = 0.441, q = 6.096 | - | - | - | Not allowed |
| M8 | 4 | −1347.154 | p0 = 0.999, p = 0.441, q = 6.096, (p1 = 0.000), ω = 1.000 | M8 vs. M7: 0.000 | 2 | 1.000 | None |
| Model | P a | LnL b | Estimates of Parameters | ΔLRT c | df | p | Positively Selected Sites |
|---|---|---|---|---|---|---|---|
| M1: Neutral | 1 | −1363.770 | p0 = 0.926, (p1 = 0.074) | - | - | - | - |
| Branch-site models | |||||||
| Model A (I) | 3 | −1361.803 | p0 = 0.929, p1 = 0.000, (p2a + p2b = 0.071), ω2 = 1.706 | Model A vs M1: 3.934 | 2 | 1.40 × 10−1 | - |
| Model A (II) | 3 | −1346.578 | p0 = 0.684, p1 = 0.018, (p2a + p2b = 0.297), ω2 = 1.053 | Model A vs M1: 34.384 | 2 | 3.40 × 10−8 | Site for foreground lineage: 4 (at p > 0.95) |
| Model A (III) | 3 | −1363.521 | p0 = 0.927, p1 = 0.057, (p2a + p2b = 0.015), ω2 = 17.935 | Model A vs M1: 0.500 | 2 | 7.78 × 10−1 | - |
| Model A (IV) | 3 | −1363.522 | p0 = 0.925, p1 = 0.064, (p2a + p2b = 0.012), ω2 = 2.630 | Model A vs M1: 0.496 | 2 | 7.81 × 10−1 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Meng, S.; Su, W.; Bao, Y.; Lu, Y.; Yin, W.; Liu, C.; Xia, X. Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress. Int. J. Mol. Sci. 2019, 20, 1425. https://doi.org/10.3390/ijms20061425
Wang D, Meng S, Su W, Bao Y, Lu Y, Yin W, Liu C, Xia X. Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress. International Journal of Molecular Sciences. 2019; 20(6):1425. https://doi.org/10.3390/ijms20061425
Chicago/Turabian StyleWang, Dongli, Sen Meng, Wanlong Su, Yu Bao, Yingying Lu, Weilun Yin, Chao Liu, and Xinli Xia. 2019. "Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress" International Journal of Molecular Sciences 20, no. 6: 1425. https://doi.org/10.3390/ijms20061425




