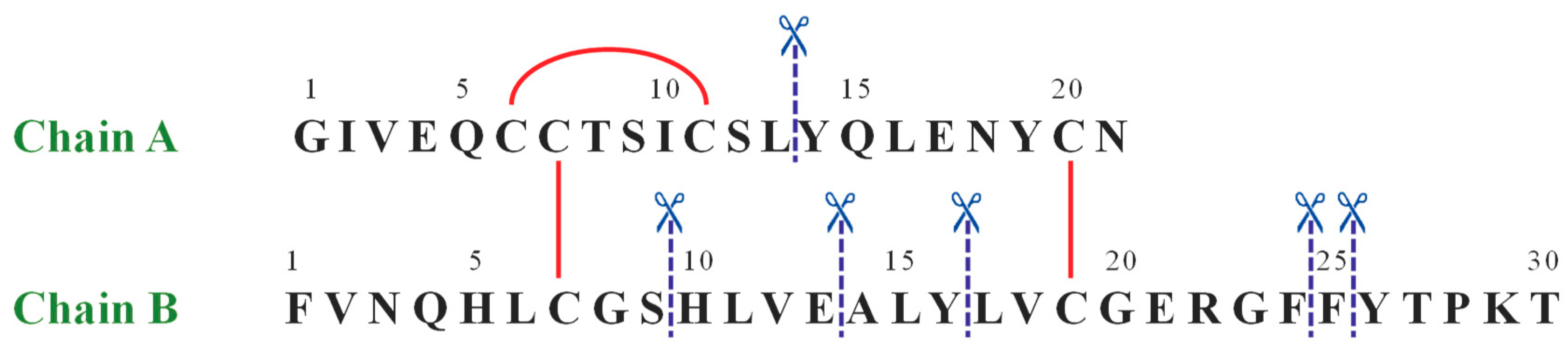Abstract
Insulin-degrading enzyme (IDE) was applied to catalyze hydrolysis of Nociceptin/Orphanin 1-16 (OFQ/N) to show the involvement of the enzyme in degradation of neuropeptides engaged in pain transmission. Moreover, IDE degradative action towards insulin (Ins) was inhibited by the OFQ/N fragments, suggesting a possible regulatory mechanism in the central nervous system. It has been found that OFQ/N and Ins affect each other degradation by IDE, although in a different manner. Indeed, while the digestion of OFQ/N is significantly affected by the presence of Ins, the kinetic profile of the Ins hydrolysis is not affected by the presence of OFQ/N. However, the main hydrolytic fragments of OFQ/N produced by IDE exert inhibitory activity towards the IDE-mediated Ins degradation. Here, we present the results indicating that, besides Ins, IDE cleaves neuropeptides and their released fragments act as inhibitors of IDE activity toward Ins. Having in mind that IDE is present in the brain, which also contains Ins receptors, it cannot be excluded that this enzyme indirectly participates in neural communication of pain signals and that neuropeptides involved in pain transmission may contribute to the regulation of IDE activity. Finally, preliminary results on the metabolism of OFQ/N, carried out in the rat spinal cord homogenate in the presence of various inhibitors specific for different classes of proteases, show that OFQ/N proteolysis in rat spinal cord could be due, besides IDE, also to a cysteine protease not yet identified.
1. Introduction
Diabetes mellitus (DM) is a disease that is spread all around the world and represents a global public health problem [1]. Two major diabetes types can develop (not considering gestational diabetes): type 1 and type 2, characterized by a lack of insulin (Ins) production and by Ins resistance, respectively. Common symptoms and problems of untreated diabetes are excessive thirst and hunger, visual disturbances, frequent urination (from urinary tract infections or kidney problems), weight loss or gain, fatigue, risk of heart disease and infections, irritability, slow-healing wounds, damaged blood vessels, and nerve damage [2,3,4]. Moreover, diabetic patients have a high pain threshold [5,6], whereas Ins has an antinociceptive effect, widely reported in the literature [7].
Nociceptin/Orphanin FN(1-17) (OFQ/N; FN denotes the presence of phenylalanine (F) at the N-terminal position and glutamine (N) at the C-terminal side) is considered to be a new member of the opioid family, showing sequence homology with the main endogenous opioid peptides of mammals, especially with dynorphin A. The primary amino acid sequences of OFQ/N and certain other endogenous peptides exhibit a common characteristic: the N-terminal tetrapeptide Phe-Gly-Gly-Phe of OFQ/N is similar to the sequence Tyr-Gly-Gly-Phe, which is present in all other opioid peptides. Despite the fact that N-terminal Tyr is essential for binding to the three known opioid receptors for dynorphins, endorphins, and enkephalins, OFQ/N binds to the NOP receptor, which belongs to the opioid receptor family and it was found that it does not bind opiates with high affinity [8]. The receptor was discovered in 1994 [9] and the NOP-OFQ/N system shows different pharmacological actions from the opioid receptor system, whereas different studies have outlined some roles of the NOP-OFQ/N system in nociception [10,11]. Although the route followed by OFQ/N in pain regulation is not perfectly clear, it is well recognized that this peptide is pain-inducing and causes hyperalgesia, as ceasing OFQ/N signaling was shown to raise pain threshold [12]. In any case, it is worth highlighting that the physiological activity of neuropeptides is regulated by the proteolytic processes that take place in the central nervous system. These enzymatic processes transform neuropeptides into shorter fragments, which can have distinct biological functions in comparison with the parent peptides from which they have been generated. In the case of OFQ/N, only very few studies investigate the metabolic processes undergone by the peptide and its fragments [13]. In this work, we report that insulin-degrading enzyme (IDE), an enzyme known for its multifaceted roles in cells [14,15,16,17] and its capability to degrade several different peptides [18,19,20,21], is capable of degrading OFQ/N in vitro, producing a characteristic proteolytic pattern. As IDE is present in the brain [22], which also contains Ins pools, it cannot be excluded that this enzyme indirectly participates in neural communication of pain signals and that neuropeptides involved in pain transmission may contribute to the regulation of IDE activity. Moreover, we have preliminary investigated the degradation of OFQ/N in the rat spinal cord, excluding the action of metalloproteases such as IDE by using EDTA, to screen for other enzymes that may also be responsible of the peptide proteolysis in vivo. Results show that, besides IDE, a cysteine protease is involved in the truncation of OFQ/N into shorter fragments, which remains to be identified.
2. Results
The IDE-mediated hydrolysis of OFQ/N in vitro yielded several peptides, which were detected and assigned by applying UPLC-HRMS. The high accuracy of the detected m/z values related to the most abundant peptides ions, coupled to their MS/MS acquisition has been used to undoubtedly identify the peptide sequence. The list of the most abundant hydrolytic peptides (Table 1) clearly shows that all the peptide bonds between Arg(8) and Lys(13) are cleaved by IDE, thus forming 1-8, 1-9, 1-10, 1-11, and 1-12 peptides. Only a N-truncated peptide (2-11) has been detected.

Table 1.
List of all the identified IDE-promoted OFQ/N hydrolytic peptides and their LC-MS features
The same approach has been used to characterize by LC-MS the IDE-induced hydrolytic pattern of Ins. In Figure 1 the amino acid sequence of human Ins is reported.
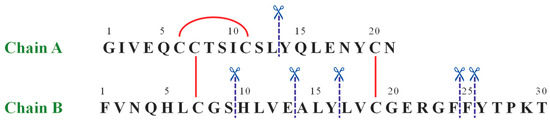
Figure 1.
The amino acid sequence of human insulin. The red straight lines show intra- and inter-chain disulfide bridges, whereas the blue dashed lines label the IDE-catalyzed cleavage sites.
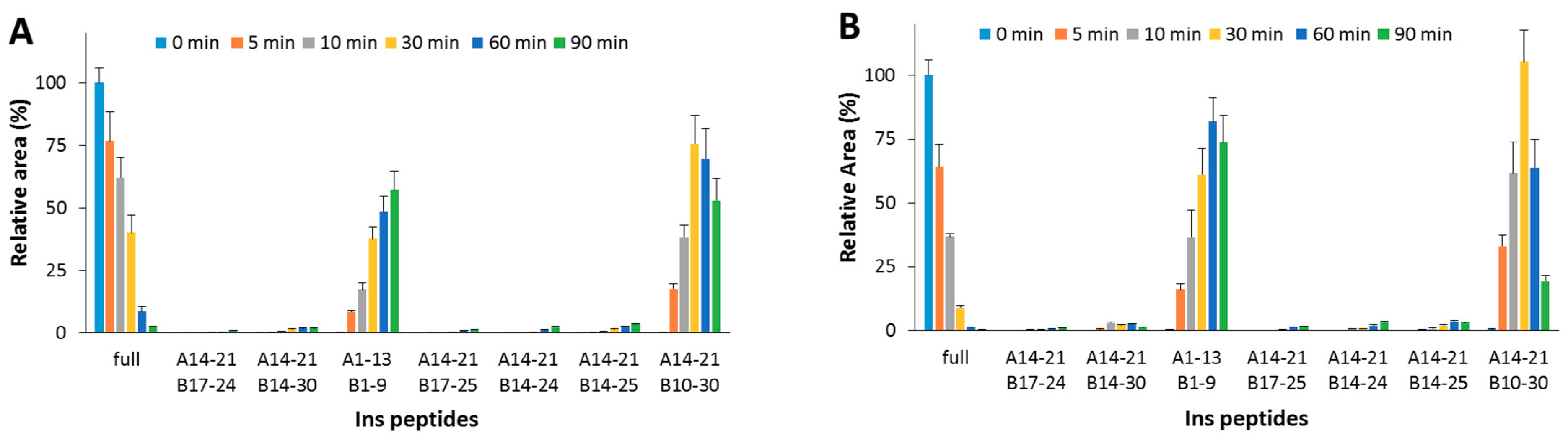
The detected peptides (Table 2 and Figure 2A) clearly confirm the attribution previously reported [23,24,25]. In the experimental conditions used, A1-13_B1-9 and A14-21_B10-30 are the main products. The content of the full-length substrate is more than halved after 30 min incubation and it almost disappears after 90 min. When OFQ/N was co-incubated with Ins and IDE (Figure 2B), the kinetic profiles of both the Ins degradation and the formation of the digested peptides did not significantly differ from those detected without OFQ/N.

Table 2.
List of all the identified IDE-promoted Ins hydrolytic peptides and their LC-MS features. The sulfur atoms involved in disulphide bonds are reported in bold and underlined (Amino acid sequence column). “A” denotes insulin chain A and “B” denotes insulin chain B.
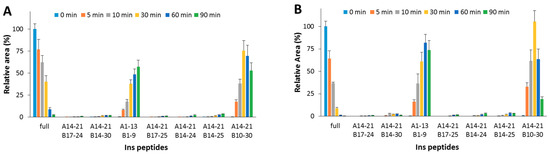
Figure 2.
Hydrolytic pattern of the time-course Ins degradation catalyzed by IDE. The content of the full-length substrate (full) and the main hydrolytic peptides was detected in the absence (A) and in the presence (B) of OFQ/N, co-incubated with Ins and IDE at 37 °C for 90 min.
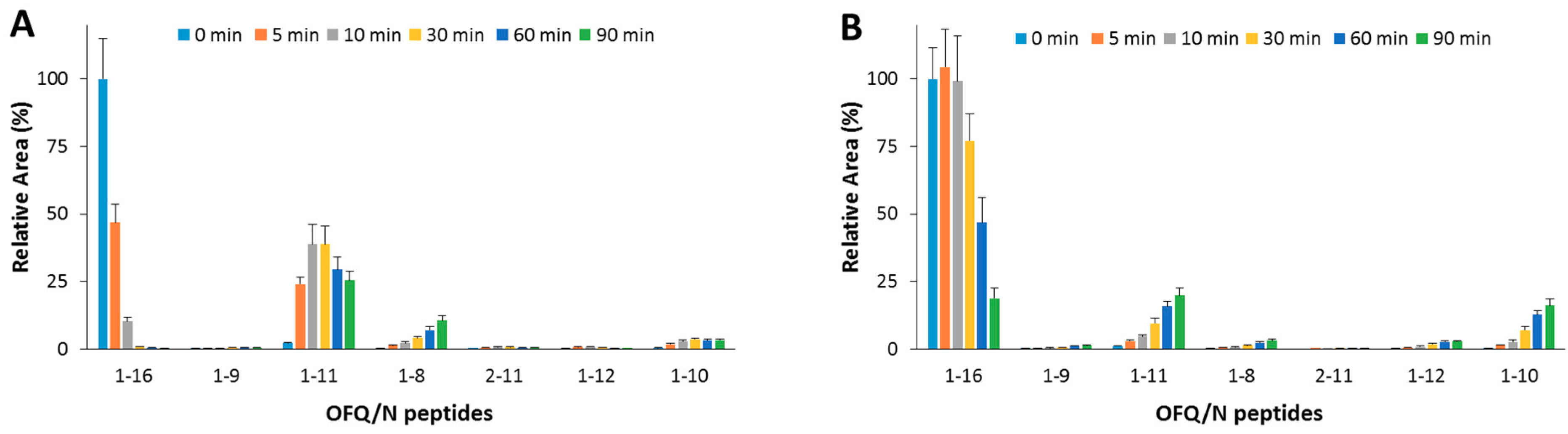
As for the IDE-mediated OFQ/N degradation (Figure 3A), the substrate was almost completely degraded within 10 min; the first and main product (1-11) reached the highest amount after 30 min; 1-8 and 1-10 are also formed from the OFQ/N as well as from the 1-11 fragment processing.
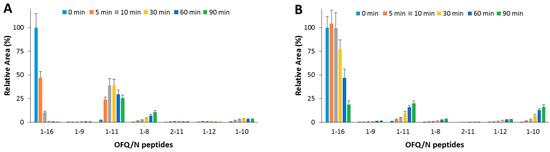
Figure 3.
Hydrolytic pattern of the time-course OFQ/N degradation catalyzed by IDE. The content of the full-length substrate (1-16) and the main hydrolytic peptides was detected in the absence (A) and in the presence (B) of Ins, co-incubated with OFQ/N and IDE at 37 °C for 90 min.
When OFQ/N and Ins (1:1 molar ratio) were co-incubated with IDE (Figure 3B), OFQ/N ended up 80% of the original value after a 30 min reaction. The 1-11 fragment was still the main product, yet formation of this peptide was inhibited by the presence of Ins. Formation of 1-8 was reduced as well. On the contrary, the content of 1-10 fragment slightly increased.
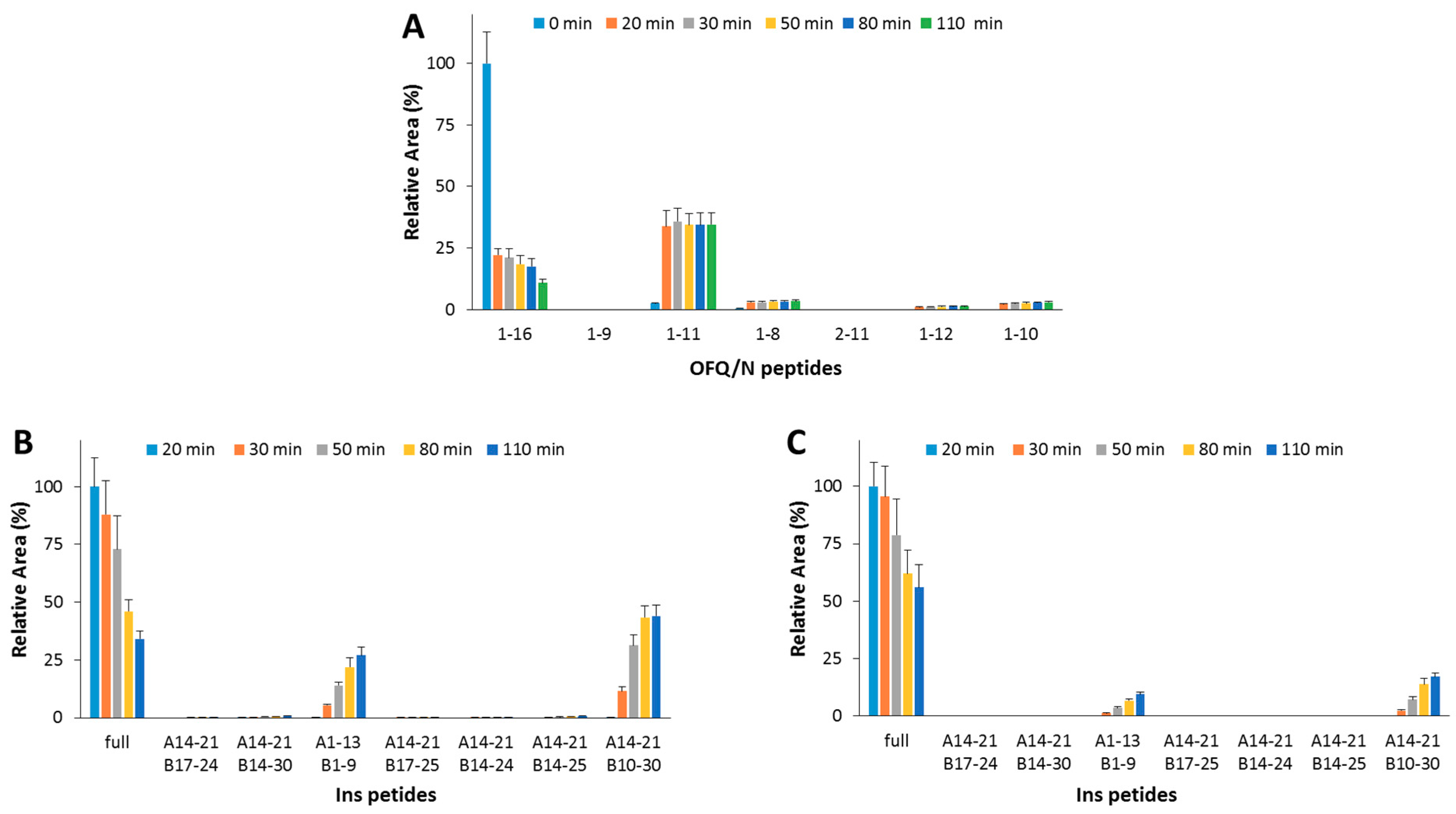
In order to assess the effect (if any) of the OFQ/N hydrolytic fragments on the IDE-catalyzed Ins cleavage, OFQ/N was pre-incubated at 37 °C with IDE. After 20 min, Ins was added to the reaction mixture. At this point, the content of OFQ/N is 5-fold lower than its starting value (Figure 4A), whereas, after the addition of Ins, the amounts of neither the substrate (1-16) nor the main hydrolytic peptides (1-11, 1-10, and 1-8) significantly change. This clearly means that IDE-mediated digestion of the residual amount of OFQ/N after the pre-incubation step is inhibited by Ins even more than that reported for the co-incubation of the two substrates (Ins and OFQ/N). Focusing on the Ins digestion, the hydrolytic pattern was analyzed when IDE was pre-incubated alone or with OFQ/N for 20 min at 37 °C. In the first case (Figure 4B), the amount of full-length Ins gradually decreases and the content of released peptides increases. If OFQ/N is pre-incubated with IDE (Figure 4C), the hydrolysis and the formation rates of the Ins fragments significantly decrease. Differently from the results obtained for the co-incubation of Ins and OFQ/N with IDE, pre-incubation of OFQ/N with IDE clearly slows down the Ins hydrolysis.
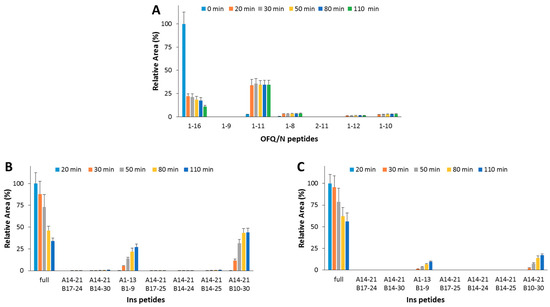
Figure 4.
Hydrolytic pattern of the time-course OFQ/N (A) degradation catalyzed by IDE at 37 °C. OFQ/N was incubated with IDE for 20 min at 37 °C before the addition of Ins. The kinetic profile of the Ins degradation was analyzed when IDE was pre-incubated alone (B) or with OFQ/N (C) at 37 °C for 20 min.
Taking into consideration the above reported results as well as the fact that IDE is present in CNS, degradation of OFQ/N in the rat spinal cord was then investigated and the same fragments found in vitro for the purified enzyme were detected. Afterwards, EDTA was added to the solution in order to verify if other proteases might also be involved in the degradation of OFQ/N in the rat spinal cord and the results are reported in Figure 5. The rationale of such study was to verify the hypothesis that processing of OFQ/N, at least partially, may depend on other proteases besides IDE.
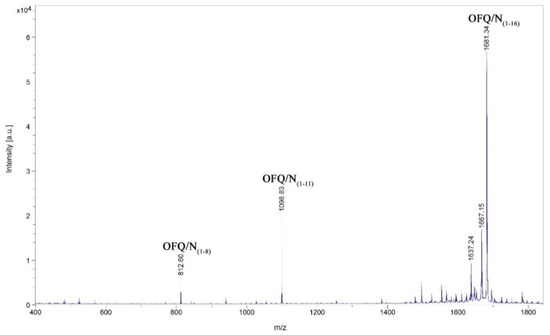
Figure 5.
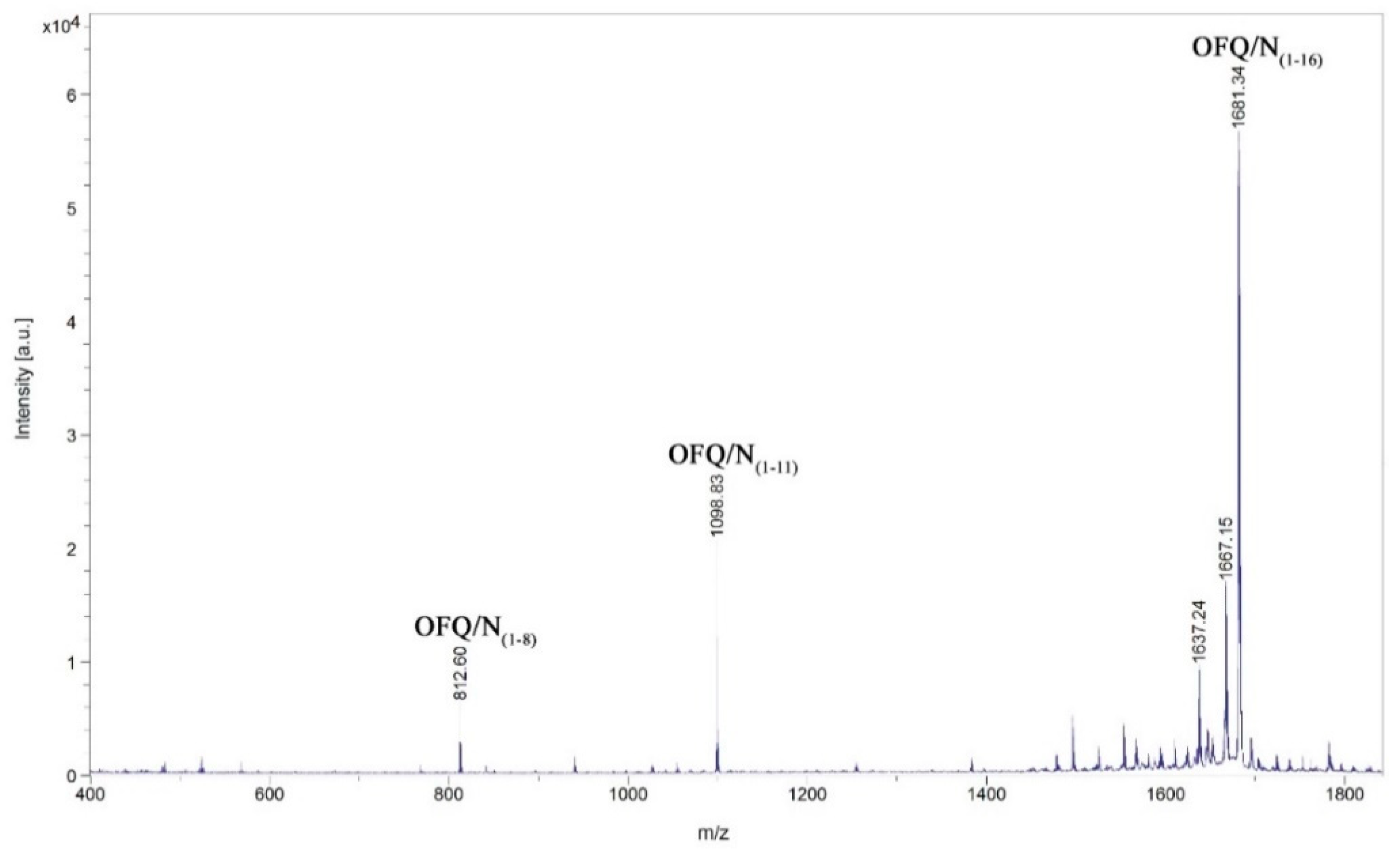
Identification by MALDI-MS of the major products after incubation of the neuropeptide OFQ/N with the homogenate obtained from the rat spinal cord. The incubation was performed using one microliter of the whole homogenate diluted with 19 µL of 20 mM Tris-HCl, 1 mM EDTA, pH 7.8, and 10 µL of Milli-Q water. The peptide concentration was 0.5 µg/mL, the incubation time was 30 min at 37 °C.
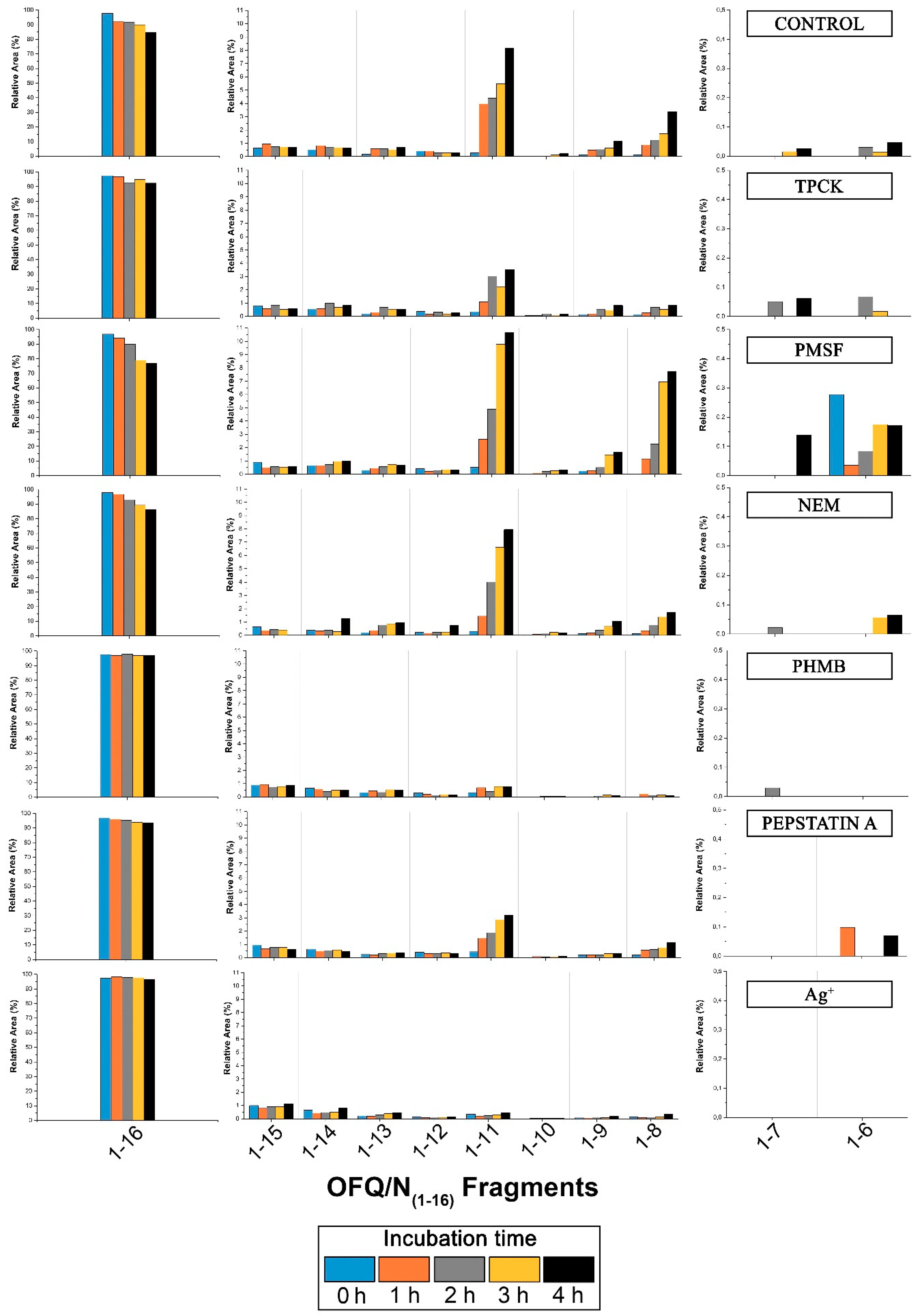
Although in some cases the intensities are rather low, we could assign signals at 1098.83, 1496.19, 1383.08, 940.72, and 812.61 m/z, corresponding to the OFQ/N fragments (1-11), (1-14), (1-13), (1-9), and (1-8), respectively. The time course of the reaction in the presence of different classes of peptidases inhibitors is reported in Figure 6. In particular, TPCK and PMSF are serine proteases inhibitors; NEM and PHMB are cysteine proteases inhibitors; Pepstatin A is aspartyl proteases inhibitor; Ag+ is a strong metalloprotease inhibitor [18,19]. In each histogram presented in Figure 6, the presence of different fragments from (1-16), the parent peptide, is clearly visible. The first fragments (from 1-15 to 1-12) are probably produced by a carboxypeptidase, an enzyme able to cleave the initial peptide, one amino acid at a time, from the C-terminal side of the chain.
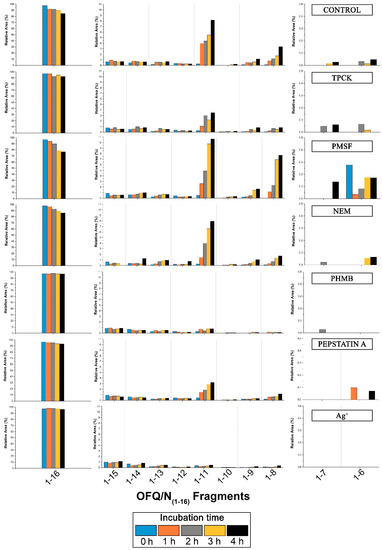
Figure 6.
Time-course of the products after cleavage by rat spinal cord peptidases with the influence of different inhibitors on the metabolism. Bar graphs are relative to all the areas under the peaks detected by MALDI. On the left and on the right part of each histogram, relative areas of OFQ/N, fragments (1-7) and (1-6) are shown on a different scale due to the different values compared to all the other fragment areas.
3. Discussion
We have found that Ins and OFQ/N compete for the interaction with IDE, thus affecting their own hydrolytic patterns. Indeed, the digestion of OFQ/N is significantly affected by the presence of Ins. On the contrary, the kinetic profile of the Ins hydrolysis does not clearly change in the presence of OFQ/N. However, the main hydrolytic peptides of OFQ/N exert an inhibitor activity towards the IDE-mediated Ins degradation and pre-incubation is necessary to compensate for the lower affinity of OFQ/N towards IDE if compared to the one between Ins and IDE.
The OFQ/N degradation in rat spinal cord has also been investigated and the most important findings concerns the (1-11), (1-7), and (1-6) fragments and their peak areas in the presence of different inhibitors, because these shorter sequences are the bioactive ones. Indeed, as reported in the literature, when intrathecally injected to mouse, (1-11) shows a naloxone-dependent analgesic action [26,27], whereas (1-7) fragment blocks hyperalgesia induced by OFQ/N injected in mice [28]. (1-11) fragment is later reduced to (1-6) fragment as a final product. The latter showed to have a bi-phasic effect in different analgesic tests: after ICV (Intracerebroventricular) and intrathecal injection it causes antinociception followed by a hyperalgesia. The cleavage of OFQ/N into the desired fragments is not efficiently inhibited by PMSF or TPCK-serine proteases inhibitors. The graph relative to the latter shows lower abundance of the (1-11) fragment compared to all the other histograms, but this could be due to the presence of methanol that stops enzyme activity in the incubation mixture. In fact, methanol was used to allow the dissolution of TPCK in water. Also, NEM does not show inhibition of the production of the desired bioactive fragments, whereas Pepstatin A slightly inhibits the formation of the (1-11) fragment. On the contrary, PHMB, a general cysteine proteases inhibitor, blocks the formation of the (1-11)fragment more than all the other inhibitors used during the experiment. This suggests that a cysteine protease is involved in the truncation of OFQ/N into shorter fragments with biological activity. Moreover, according to the data presented in Figure 6, PHMB inhibits the formation of (1-7) and (1-6) too.
4. Materials and Methods
IDE degradation assays: Insulin (Ins, 2 µM, purchased from Sigma, Milan, Italy) and nociceptin peptide (OFQ/N, 2 µM, synthesized by the Fmoc solid phase synthesis and purified in our laboratory using RP HPLC) were tested for their purity using electrospray mass spectrometry (over 98% purity), and hydrolyzed in the presence of insulin-degrading enzyme (IDE, 30 nM, purchased from Giotto Biotech, Florence, Italy) in phosphate buffer 1 mM (pH 7.4, Sigma). We applied 1-16 sequence of OFQ/N because there is no difference in binding of this fragment to NOP receptors. Ins and OFQ/N were hydrolyzed both separately and in a mixture. Moreover, a pre-incubation of OFQ/N with IDE for 20 min was also performed before the addition of Ins. In order to quench the reaction, the samples were diluted 1:1 with 1% TFA aqueous solution.
The peptide content of all samples was analyzed using a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Milan, Italy) coupled to an Ultimate 3000 HPLC RSLCnano system (Dionex Thermo Scientific, Milan, Italy) through an EASY-Spray source (Thermo Scientific, Milan, Italy) by using the instrument settings previously reported [29].
Tissue homogenate preparation: One rat spinal cord tissue was homogenized in 1 mL of ice-cold 20 mM Tris-HCl (Sigma, Milan, Italy) and 1 mM EDTA (Sigma, Milan, Sigma) using mechanical homogenizer. The obtained homogenate was centrifuged at 5000× g for 15 min at 4 °C. The supernatant was then centrifuged at 20,000× g for 15 min at 4 °C. Finally, the supernatant was divided into small aliquots and kept frozen at −80 °C [13].
Tissue extract (supernatant) incubation assays: Incubations were performed using one microliter of tissue extract diluted with 19 µL of 20 mM Tris-HCl 1 mM EDTA buffer pH 7.8 and 10 µL of Milli-Q water. Then, the reaction was initiated by the addition of 10 µL of OFQ/N 0.3 mM and continued at 37 °C for 30 min. Then, 40 µL of ice-cold methanol was used to stop the reaction. Incubation was repeated as mentioned above but adding different peptidase inhibitors. The inhibitors used for the incubation were: TPCK (tosyl phenylalanyl chloromethyl ketone) 1 mM, PMSF (phenylmethylsulfonyl fluoride) 1 mM, NEM (N-ethylmaleimide) 1 mM, PHMB (4-(hydroxymercuri)benzoic acid) 0.25 mM, pepstatin A 0.01 mM, and Ag+ 1 mM. Enzyme assay including spinal cord homogenate was also validated using dynorphin B, as described previously [30]. One microliter aliquots taken from each incubation mixture were analyzed using MALDI mass spectrometry. α-Cyano-4-hydroxycinnamic acid (4-HCCA) was used as matrix, and the final samples were spotted onto AnchorChip™ plate (Bruker Daltonics, 2013). The standard used for the calibration of the instrument is the Peptide Calibration Standard II by Bruker Daltonics, Macerata, Italy. Details and parameters of the instrument set-up were: mass range: 340–4000 Da; detector gain/reflector: 6.3x (2480 V); sample rate and digitizer settings: 2.50 GS/s; smartbeam parameter set: 4_large; positive ion mode; matrix suppression: off; random walk: complete sample; shots at raster spot: 200; limit diameter to: 800 µm; laser power: 20%; shots: 5000.
5. Conclusions
We have investigated the degradation of the OFQ/N peptide in vitro by purified IDE. Additionally, we performed a preliminary screen for other peptidases potentially cleaving OFQ/N in the rat spinal cord. We have found that IDE is capable of degrading the neuropeptide to generate shorter fragments that are reported to exhibit an anti-nociceptive effect. We have shown that, in vitro, Ins slows down the degradation of the OFQ/N peptide by IDE, whereas the OFQ/N peptide does not affect Ins degradation by IDE. However, the OFQ/N smaller fragments produced by the action of IDE on the OFQ/N peptide affects Ins proteolysis by IDE, as demonstrated by pre-incubating the enzyme with the OFQ/N peptide before Ins addition. These results demonstrate that Ins dyshomeostasis due to the type 2 or type 1 diabetes can have a direct impact on pain transmission and pain threshold through an IDE mediated mechanism. This result is in line with all the recent findings, which demonstrate a multifaceted role for this enzyme in the brain [14,15,16,31].
Moreover, preliminary results with the use of rat spinal cord indicate that a cysteine protease, besides IDE, could be involved in the truncation of OFQ/N into shorter fragments in vitro.
Future experiments, carried out in the presence of specific IDE inhibitors and Ins fragments, will allow assessment of the relevance of our findings in vivo, thus elucidating whether nociceptin degradation by IDE is the main process responsible for the reported higher pain threshold in diabetic patients or if other biochemical pathways are also involved [32].
Author Contributions
Conceptualization, G.G. and J.S.; methodology, G.G., J.S., P.M.; software, G.A.Z., F.B., P.M.; validation, G.A.Z., F.B., P.M. and I.M.M.A.; formal analysis, F.B., P.M., J.S., I.M.M.A. and G.G.; investigation, G.A.Z. and I.M.M.A.; resources, F.B., G.G. and J.S.; data curation, F.B., P.M., J.S., I.M.M.A. and G.G.; writing—original draft preparation, G.G., J.S.; writing—review and editing, G.G. and J.S.; visualization, G.G. and J.S.; supervision, F.B., P.M., G.G. and J.S.; project administration, G.G. and J.S.; funding acquisition, F.B., G.G. and J.S.”.
Funding
The authors thank the INCIPIT-COFUND project co-funded by HORIZON 2020/Marie Sklodowska Curie Actions and MIUR (PRIN Prot. 20157WZM8A) for the financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaul, K.; Tarr, J.M.; Ahmad, S.I.; Kohner, E.M.; Chibber, R. Introduction to diabetes mellitus. Adv. Exp. Med. Biol. 2012, 771, 1–11. [Google Scholar] [PubMed]
- Saidian, M.; Lakey, J.R.T.; Ponticorvo, A.; Rowland, R.; Baldado, M.; Williams, J.; Pronda, M.; Alexander, M.; Flores, A.; Shiri, L.; et al. Characterisation of impaired wound healing in a preclinical model of induced diabetes using wide-field imaging and conventional immunohistochemistry assays. Int. Wound J. 2019, 16, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Sharma, A.K.; Nag, D.; Das, A.; Datta, S.; Ganguli, A.; Goel, V.; Rajput, S.; Chakrabarti, G.; Basu, B.; et al. Novel nano-insulin formulation modulates cytokine secretion and remodeling to accelerate diabetic wound healing. Nanomedicine 2019, 15, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Masood, N.; Ahmed, R.; Tariq, M.; Ahmed, Z.; Masoud, M.S.; Ali, I.; Asghar, R.; Andleeb, A.; Hasan, A. Silver nanoparticle impregnated chitosan-PEG hydrogel enhances wound healing in diabetes induced rabbits. Int. J. Pharm. 2019, 559, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Telli, O.; Cavlak, U. Measuring the pain threshold and tolerance using electrical stimulation in patients with Type II diabetes mellitus. J. Diabetes Complicat. 2006, 20, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Kukidome, D.; Nishikawa, T.; Sato, M.; Igata, M.; Kawashima, J.; Shimoda, S.; Matsui, K.; Obayashi, K.; Ando, Y.; Araki, E. Measurement of small fibre pain threshold values for the early detection of diabetic polyneuropathy. Diabet. Med. 2016, 33, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Yamaguchi, I. Insulin attenuates formalin-induced nociceptive response in mice through a mechanism that is deranged by diabetes mellitus. J. Pharmacol. Exp. Ther. 1997, 281, 315–321. [Google Scholar]
- Mollereau, C.; Parmentier, M.; Mailleux, P.; Butour, J.L.; Moisand, C.; Chalon, P.; Caput, D.; Vassart, G.; Meunier, J.C. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett. 1994, 341, 33–38. [Google Scholar] [CrossRef]
- Bunzow, J.R.; Saez, C.; Mortrud, M.; Bouvier, C.; Williams, J.T.; Low, M.; Grandy, D.K. Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, delta or kappa opioid receptor type. FEBS Lett. 1994, 347, 284–288. [Google Scholar] [CrossRef]
- Calo’, G.; Guerrini, R.; Rizzi, A.; Salvadori, S.; Regoli, D. Pharmacology of nociceptin and its receptor: a novel therapeutic target. Br. J. Pharmacol. 2000, 129, 1261–1283. [Google Scholar] [CrossRef]
- Calo’, G.; Rizzi, A.; Bigoni, R.; Guerrini, R.; Salvadori, S.; Regoli, D. Pharmacological profile of Nociceptin/Orphanin FQ receptors. Clin. Exp. Pharmacol. Physiol. 2002, 29, 223–228. [Google Scholar] [CrossRef]
- Meunier, J.C.; Mollereau, C.; Toll, L.; Suaudeau, C.; Moisand, C.; Alvinerie, P.; Butour, J.L.; Guillemot, J.C.; Ferrara, P.; Monsarrat, B. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature 1995, 377, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Suder, P.; Kotlinska, J.; Smoluch, M.T.; Sällberg, M.; Silberring, J. Metabolic fate of nociceptin/orphanin FQ in the rat spinal cord and biological activity of its released fragment. Peptides 1999, 20, 239–247. [Google Scholar] [CrossRef]
- Grasso, G.; Lanza, V.; Malgieri, G.; Fattorusso, R.; Pietropaolo, A.; Rizzarelli, E.; Milardi, D. The insulin degrading enzyme activates ubiquitin and promotes the formation of K48 and K63 diubiquitin. Chem. Commun. (Camb) 2015, 51, 15724–15727. [Google Scholar] [CrossRef] [PubMed]
- Sbardella, D.; Tundo, G.R.; Coletta, A.; Marcoux, J.; Koufogeorgou, E.I.; Ciaccio, C.; Santoro, A.M.; Milardi, D.; Grasso, G.; Cozza, P.; et al. The insulin-degrading enzyme is an allosteric modulator of the 20S proteasome and a potential competitor of the 19S. Cell Mol. Life Sci. 2018, 75, 3441–3456. [Google Scholar] [CrossRef] [PubMed]
- Bellia, F.; Lanza, V.; Ahmed, I.M.M.; Garcia-Vinuales, S.; Veiss, E.; Arizzi, M.; Calcagno, D.; Milardi, D.; Grasso, G. Site directed mutagenesis of insulin-degrading enzyme allows singling out the molecular basis of peptidase versus E1-like activity: the role of metal ions. Metallomics 2019, 11, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Sbardella, D.; Ciaccio, C.; Grasso, G.; Gioia, M.; Coletta, A.; Polticelli, F.; Di Pierro, D.; Milardi, D.; Van Endert, P.; et al. Multiple functions of insulin-degrading enzyme: a metabolic crosslight? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 554–582. [Google Scholar] [CrossRef]
- Grasso, G.; Pietropaolo, A.; Spoto, G.; Pappalardo, G.; Tundo, G.R.; Ciaccio, C.; Coletta, M.; Rizzarelli, E. Copper(I) and Copper(II) Inhibit Aβ Peptides Proteolysis by Insulin-Degrading Enzyme Differently: Implications for Metallostasis Alteration in Alzheimer’s Disease. Chem. Eur. J. 2011, 17, 2752–2762. [Google Scholar] [CrossRef]
- Grasso, G.; Salomone, F.; Tundo, G.R.; Pappalardo, G.; Ciaccio, C.; Spoto, G.; Pietropaolo, A.; Coletta, M.; Rizzarelli, E. Metal ions affect insulin-degrading enzyme activity. J. Inorg. Biochem. 2012, 117, 351–358. [Google Scholar] [CrossRef][Green Version]
- Bellia, F.; Grasso, G. The role of copper(II) and zinc(II) in the degradation of human and murine IAPP by insulin-degrading enzyme. J. Mass. Spectrom. 2014, 49, 274–279. [Google Scholar] [CrossRef]
- Ciaccio, C.; Tundo, G.R.; Grasso, G.; Spoto, G.; Marasco, D.; Ruvo, M.; Gioia, M.; Rizzarelli, E.; Coletta, M. Somatostatin: a novel substrate and a modulator of insulin-degrading enzyme activity. J. Mol. Biol. 2009, 385, 1556–1567. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.M.; Barrett, E.J. Insulin transport into the brain. Am. J. Physiol. Cell Physiol. 2018, 315, C125–C136. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Rizzarelli, E.; Spoto, G. AP/MALDI-MS complete characterization of the proteolytic fragments produced by the interaction of insulin degrading enzyme with bovine insulin. J. Mass. Spectrom. 2007, 42, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Rizzarelli, E.; Spoto, G. The proteolytic activity of insulin-degrading enzyme: a mass spectrometry study. J. Mass. Spectrom. 2009, 44, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Bellia, F.; Pietropaolo, A.; Grasso, G. Formation of insulin fragments by insulin-degrading enzyme: the role of zinc(II) and cystine bridges. J. Mass. Spectrom. 2013, 48, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.C.; Leventhal, L.; Bolan, E.; Pasternak, G.W. Pharmacological characterization of orphanin FQ/nociceptin and its fragments. J. Pharmacol. Exp. Ther. 1997, 282, 858–865. [Google Scholar] [PubMed]
- Rossi, G.C.; Perlmutter, M.; Leventhal, L.; Talatti, A.; Pasternak, G.W. Orphanin FQ/nociceptin analgesia in the rat. Brain Res. 1998, 792, 327–330. [Google Scholar] [CrossRef]
- Sakurada, C.; Sakurada, S.; Katsuyama, S.; Sasaki, J.; Tan-No, K.; Sakurada, T. Involvement of tachykinin NK1 receptors in nociceptin-induced hyperalgesia in mice. Brain Res. 1999, 841, 85–92. [Google Scholar] [CrossRef]
- Bellia, F.; Lanza, V.; Garcia-Vinuales, S.; Ahmed, I.M.M.; Pietropaolo, A.; Iacobucci, C.; Malgieri, G.; D’Abrosca, G.; Fattorusso, R.; Nicoletti, V.G.; et al. Ubiquitin binds the amyloid β peptide and interferes with its clearance pathways. Chem. Sci. 2019, 10, 2732. [Google Scholar] [CrossRef] [PubMed]
- Silberring, J.; Nyberg, F. A novel bovine spinal cord endoprotease with high specificity for dynorphin B. J. Biol. Chem. 1989, 264, 11082–11086. [Google Scholar] [PubMed]
- Grasso, G.; Mielczarek, P.; Niedziolka, M.; Silberring, J. Metabolism of Cryptic Peptides Derived from Neuropeptide FF Precursors: The Involvement of Insulin-Degrading Enzyme. Int. J. Mol. Sci. 2014, 15, 16787–16799. [Google Scholar] [CrossRef] [PubMed]
- Madiraju, S.R.M.; Poitout, V. GPCRs and Insulin Secretion: 119 and Counting. Endocrinology 2007, 148, 2598–2600. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).