Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera)
Abstract
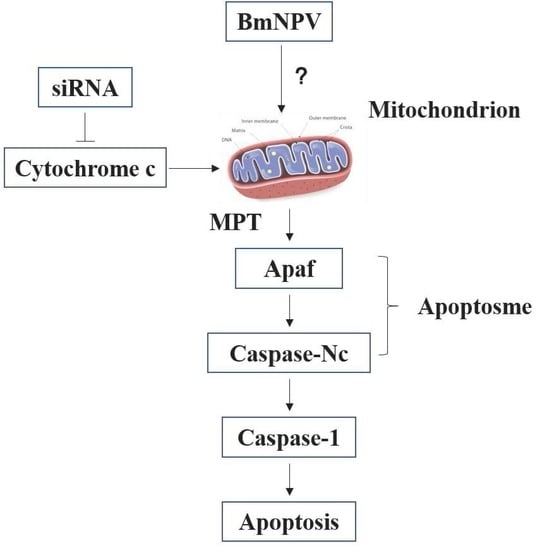
:1. Introduction
2. Results
2.1. Characterization of the Bmcytc Sequence
2.2. The Spatio-Temporal Expression Pattern of BMCYTC
2.3. Bmcytc Showed Significant Response to BmNPV Infection in Different Tissues
2.4. Selected Downstream Genes Were Downregulated after Knockdown of BMCYTC in BmN Cells
2.5. Knockdown of Bmcytc Promotes BmNPV Infection in BmN Cells
2.6. Overexpression of Bmcytc Inhibits BmNPV Early Infection in BmN Cells
2.7. Overexpression of Bmcytc Influences the Expression of Selected Downstream Genes in BmN Cells
2.8. Analysis of Apoptosis In Vivo and In Vitro
3. Discussion
4. Materials and Methods
4.1. Silkworm and BmNPV
4.2. Bioinformatics Analysis
4.3. Sample Preparation, RNA Extraction and cDNA Synthesis
4.4. Quantitative Reverse Transcription PCR (RT-qPCR)
4.5. Synthesis of siRNA
4.6. Construction of pIZT/V5-His-mCherry-Bmcytc Overexpression Vector
4.7. BmN Cell Culture and Transfection
4.8. Inhibition and Induction of Apoptosis
4.9. Genomic DNA Extraction
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| B. mori | Bombyx mori |
| Cytc | Cytochrome c |
| BmNPV | Bombyx mori nucleopolyhedrovirus |
| PCD | Programmed cell death |
| Sf9 | Spodoptera frugiperda |
| AfMNPV | Anagrapha falcifera Multiple Nuclear Polyhedrosis Virus |
| BV-EGFP | Budded virus containing EGFP-tagged |
| RT-qPCR | Quantitative reverse transcription PCR |
| BmGAPDH | B. mori glyceraldehyde-3-phosphate dehydrogenase |
| FBS | Fetal bovine serum |
| ORF | Open reading fragment |
| GFP | Green fluorescent protein |
| Apaf | Apoptotic Protease-Activating Factor |
References
- Bao, Y.Y.; Tang, X.D.; Lv, Z.Y.; Wang, X.Y.; Tian, C.H.; Xu, Y.P.; Zhang, C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [PubMed]
- Shao, Q.M.; Yang, B.; Xu, Q.Y.; Li, X.Q.; Lu, Z.Q.; Wang, C.S.; Huang, Y.P.; Soderhall, K.; Ling, E.J. Hindgut innate immunity and regulation of fecal microbiota through melanization in insects. J. Biol. Chem. 2012, 287, 14270–14279. [Google Scholar]
- Smith, C.A.; Williams, G.T.; Kingston, R.; Jenkinson, E.J.; Owen, J.J.T. Apoptosis. Nature 1989, 338, 10. [Google Scholar] [PubMed]
- Mohamad, N.; Gutiérrez, A.; Núñez, M.; Cocca, C.; Martín, G.; Cricco, G.; Medina, V.; Rivera, E.; Bergoc, R. Mitochondrial apoptotic pathways. Biocell 2005, 29, 149–161. [Google Scholar]
- Kvansakul, M. Viral infection and apoptosis. Viruses 2017, 9, 356. [Google Scholar]
- Liu, L.; Peng, J.; Liu, K.; Yang, H.; Li, Y.; Hong, H. Influence of cytochrome c on apoptosis induced by Anagrapha (Syngrapha) falcifera multiple nuclear polyhedrosis virus (AfMNPV) in insect Spodoptera litura cells. Cell Biol. Int. 2007, 31, 996–1001. [Google Scholar]
- Arnoult, D.; Parone, P.; Martinou, J.C.; Antonsson, B.; Estaquier, J.; Ameisen, J.C. Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J. Cell Biol. 2002, 159, 923. [Google Scholar]
- Sato, M.; Matsushima, K.; Kawanami, H.; Ikuhsima, Y. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 2004, 167, 405–410. [Google Scholar]
- Shan, S.; Liu, K.; Peng, J.; Yao, H.; Li, Y.; Hong, H. Mitochondria are involved in apoptosis induced by ultraviolet radiation in lepidopteran Spodoptera fitura cell line. Insect Sci. 2010, 16, 485–491. [Google Scholar]
- Huang, J.F.; Tian, M.; Lv, C.J.; Li, H.Y.; Muhammad, R.U.H.; Zhong, G.H. Preliminary studies on induction of apoptosis by abamectin in Spodoptera frugiperda (Sf9) cell line. Pestic. Biochem. Phys. 2011, 99, 256–263. [Google Scholar] [CrossRef]
- Liu, K.; Shu, D.; Song, N.; Gai, Z.; Yuan, Y.; Li, J.; Li, M.; Guo, S.; Peng, J.; Hong, H. The role of cytochrome c on apoptosis induced by Anagrapha falcifera multiple nuclear polyhedrosis virus in insect Spodoptera litura cells. PloS ONE 2012, 7, e40877. [Google Scholar]
- Carthy, C.M.; Bobby, Y.; Honglin, L.; Granville, D.J.; Decheng, Y.; Paul, C.; Caroline, C.; Mitra, E.; Rudin, C.M.; Thompson, C.B. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 2016, 313, 147–157. [Google Scholar] [CrossRef]
- Machida, K.; Tsukiyama-Kohara, K.; Seike, E.; Toné, S.; Shibasaki, F.; Shimizu, M.; Takahashi, H.; Hayashi, Y.; Funata, N.; Taya, C. Inhibition of cytochrome c release in Fas-mediated signaling pathway in transgenic mice induced to express hepatitis C viral proteins. J. Biol. Chem. 2001, 276, 12140–12146. [Google Scholar] [PubMed]
- Wang, X.Y.; Yu, H.Z.; Geng, L.; Xu, J.P.; Yu, D.; Zhang, S.Z.; Ma, Y.; Fei, D.Q. Comparative transcriptome analysis of Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic resistant strains. PloS ONE 2016, 11, e0155341. [Google Scholar]
- Wang, X.Y.; Yu, H.Z.; Xu, J.P.; Zhang, S.Z.; Yu, D.; Liu, M.H. Comparative subcellular proteomics analysis of susceptible and near-isogenic resistant Bombyx mori (Lepidoptera) larval midgut response to BmNPV infection. Sci. Rep. 2017, 7, 45690. [Google Scholar] [CrossRef]
- Wang, X.Y.; Shao, Z.M.; Zhang, Y.J.; Vu, T.T.; Wu, Y.C.; Xu, J.P.; Deng, M.J. A 1H-NMR based study of hemolymph metabonomics in different resistant silkworms, Bombyx mori (Lepidotera), after BmNPV inoculation. J. Insect Physiol. 2019, 4, 117. [Google Scholar]
- Everett, H.; Mcfadden, G. Apoptosis: An innate immune response to virus infection. Trends Microbiol. 1999, 7, 160–165. [Google Scholar]
- Luo, X.; Yanjun, L.; Zuodong, Q.; Zhiyuan, J.; Liting, X.; Yang, L.; Linyan, C.; Fulin, H.; Xiaohong, G.; Xiaoping, O. Studies on the antibacterial activity and mechanism of antimicrobial peptides against drug-resistant bacteria. J. Biomed. Nanotechnol. 2018, 14, 601. [Google Scholar]
- La, W.; Qin, X.; Xiao-Lin, Z.; Yan, Z.; Zhan-Qi, D.; Peng, C.; Min-Hui, P.; Cheng, L. Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) induces host cell autophagy to benefit infection. Viruses 2017, 10, 14. [Google Scholar]
- Wang, X.Y.; Shao, Z.M.; Chen, Q.Y.; Xu, J.P.; Sun, X.; Xu, Z.P.; Li, M.W.; Wu, Y.C. Knockdown of BmTCP-1beta delays BmNPV infection in vitro. Front. Microbiol. 2019, 10, 578. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [PubMed]
- Guo, H.; Jiang, L.; Xia, Q. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genom. 2015, 291, 999–1004. [Google Scholar] [CrossRef] [PubMed]







| Gene Name | Forward Primers (5′-3’) | Revers Primers (5′-3’) |
|---|---|---|
| Bmcytc | TCATACTCCGATGCCAATAAAGC | CATTTGCCTTCTTGAGTCCAGC |
| BmApaf | TCACAACCCTCTAAAATCACACCAG | CGACAGCCAGTAATGGGTGTATGAG |
| BmCaspase-Nc | GAGGACGATGTGAGCAGGGAT | TTCAGCAGGAACGAAATGTAGC |
| BmCaspase-1 | AACGGCAACGAAGACGAAGG | GGTGCCCGTGCGAGATTTTA |
| BmGAPDH | CGATTCAACATTCCAGAGCA | GAACACCATAGCAAGCACGAC |
| VP39 | CAACTTTTTGCGAAACGACTT | GGCTACACCTCCACTTGCTT |
| Bmcytc KX | GGGGTACCATGGGTGTACCTGCAGGAAA | GCTCTAGACTTGGTAGCAGATTTGAGATAGG |
| Primer Names | Sequences (5′-3′) |
|---|---|
| Bmcyto-1 Olig-1 | GATCACTAATACGACTCACTATAGGGGGACCGAATCTACATGGATTT |
| Bmcyto-1 Olig-2 | AAATCCATGTAGATTCGGTCCCCCTATAGTGAGTCGTATTAGTGATC |
| Bmcyto-1 Olig-3 | AAGGACCGAATCTACATGGATCCCTATAGTGAGTCGTATTAGTGATC |
| Bmcyto-1 Olig-4 | GATCACTAATACGACTCACTATAGGGATCCATGTAGATTCGGTCCTT |
| Bmcyto-2 Olig-1 | GATCACTAATACGACTCACTATAGGGCCTTATTGCCTATCTCAAATT |
| Bmcyto-2 Olig-2 | AATTTGAGATAGGCAATAAGGCCCTATAGTGAGTCGTATTAGTGATC |
| Bmcyto-2 Olig-3 | AACCTTATTGCCTATCTCAAACCCTATAGTGAGTCGTATTAGTGATC |
| Bmcyto-2 Olig-4 | GATCACTAATACGACTCACTATAGGGTTTGAGATAGGCAATAAGGTT |
| GFP Olig-1 | GATCACTAATACGACTCACTATAGGGGGAGTTGTCCCAATTCTTGTT |
| GFP Olig-2 | AACAAGAATTGGGACAACTCCCCCTATAGTGAGTCGTATTAGTGATC |
| GFP Olig-3 | AAGGAGTTGTCCCAATTCTTGCCCTATAGTGAGTCGTATTAGTGATC |
| GFP Olig-4 | GATCACTAATACGACTCACTATAGGGCAAGAATTGGGACAACTCCTT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.-Y.; Wu, K.-H.; Pang, H.-L.; Xu, P.-Z.; Li, M.-W.; Zhang, G.-Z. Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera). Int. J. Mol. Sci. 2019, 20, 4325. https://doi.org/10.3390/ijms20184325
Wang X-Y, Wu K-H, Pang H-L, Xu P-Z, Li M-W, Zhang G-Z. Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera). International Journal of Molecular Sciences. 2019; 20(18):4325. https://doi.org/10.3390/ijms20184325
Chicago/Turabian StyleWang, Xue-Yang, Kang-Hui Wu, Hui-Lin Pang, Ping-Zhen Xu, Mu-Wang Li, and Guo-Zheng Zhang. 2019. "Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera)" International Journal of Molecular Sciences 20, no. 18: 4325. https://doi.org/10.3390/ijms20184325
APA StyleWang, X.-Y., Wu, K.-H., Pang, H.-L., Xu, P.-Z., Li, M.-W., & Zhang, G.-Z. (2019). Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera). International Journal of Molecular Sciences, 20(18), 4325. https://doi.org/10.3390/ijms20184325