Identification and in Silico Characterization of GT Factors Involved in Phytohormone and Abiotic Stresses Responses in Brachypodium distachyon
Abstract
1. Introduction
2. Results
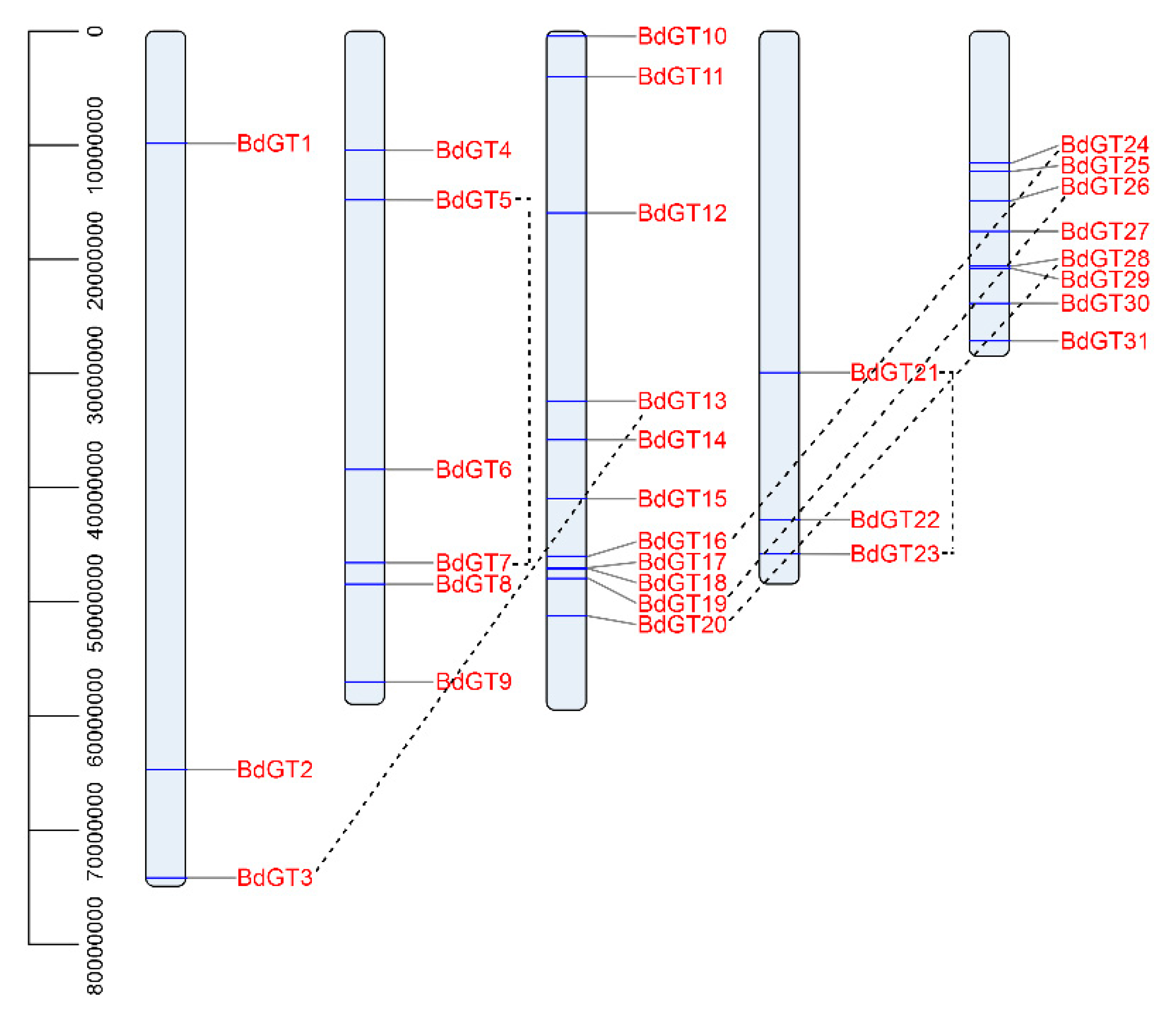
2.1. Isolation and Genomic Distribution
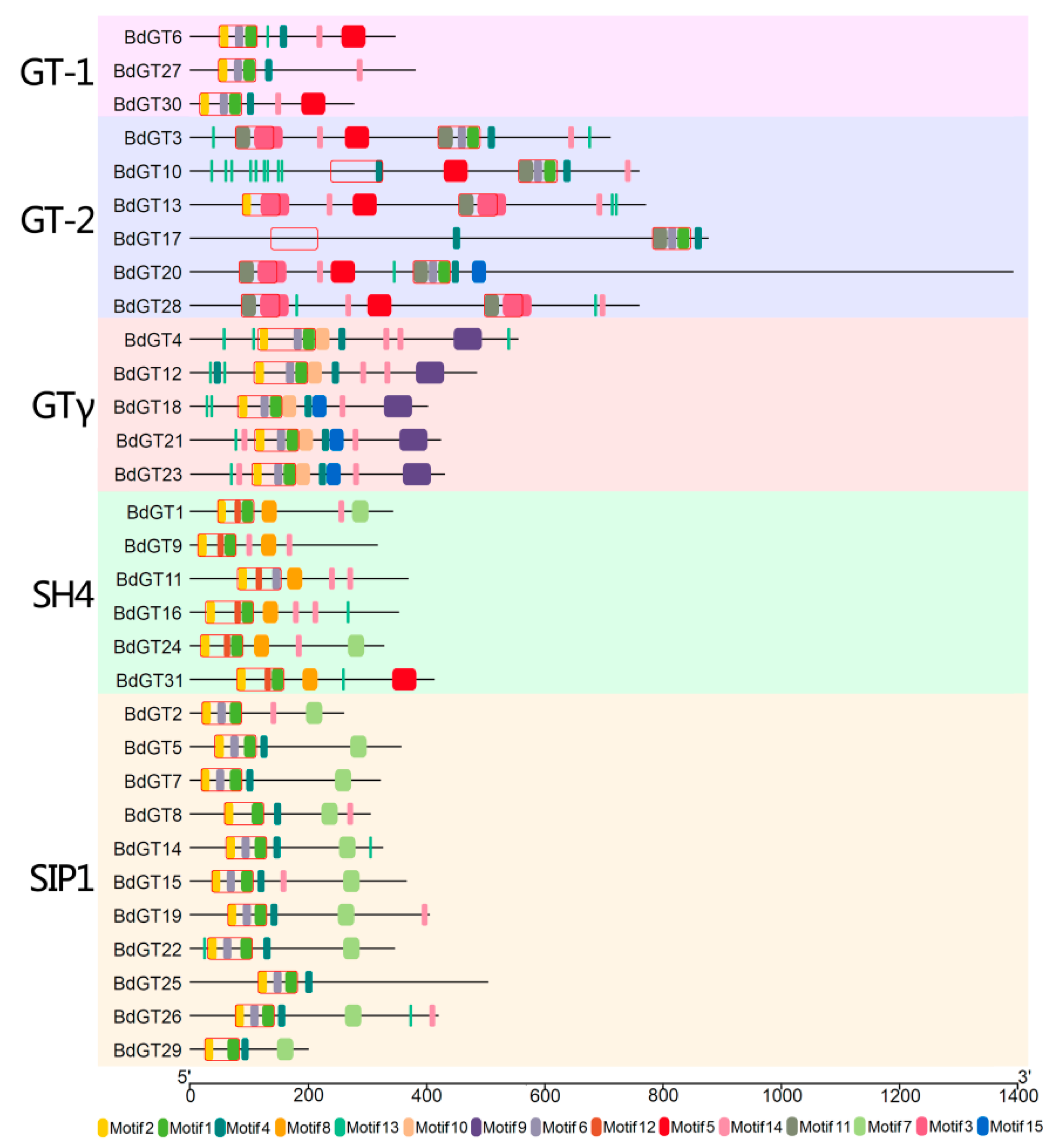
2.2. Structural Divergence Among the BdGTs
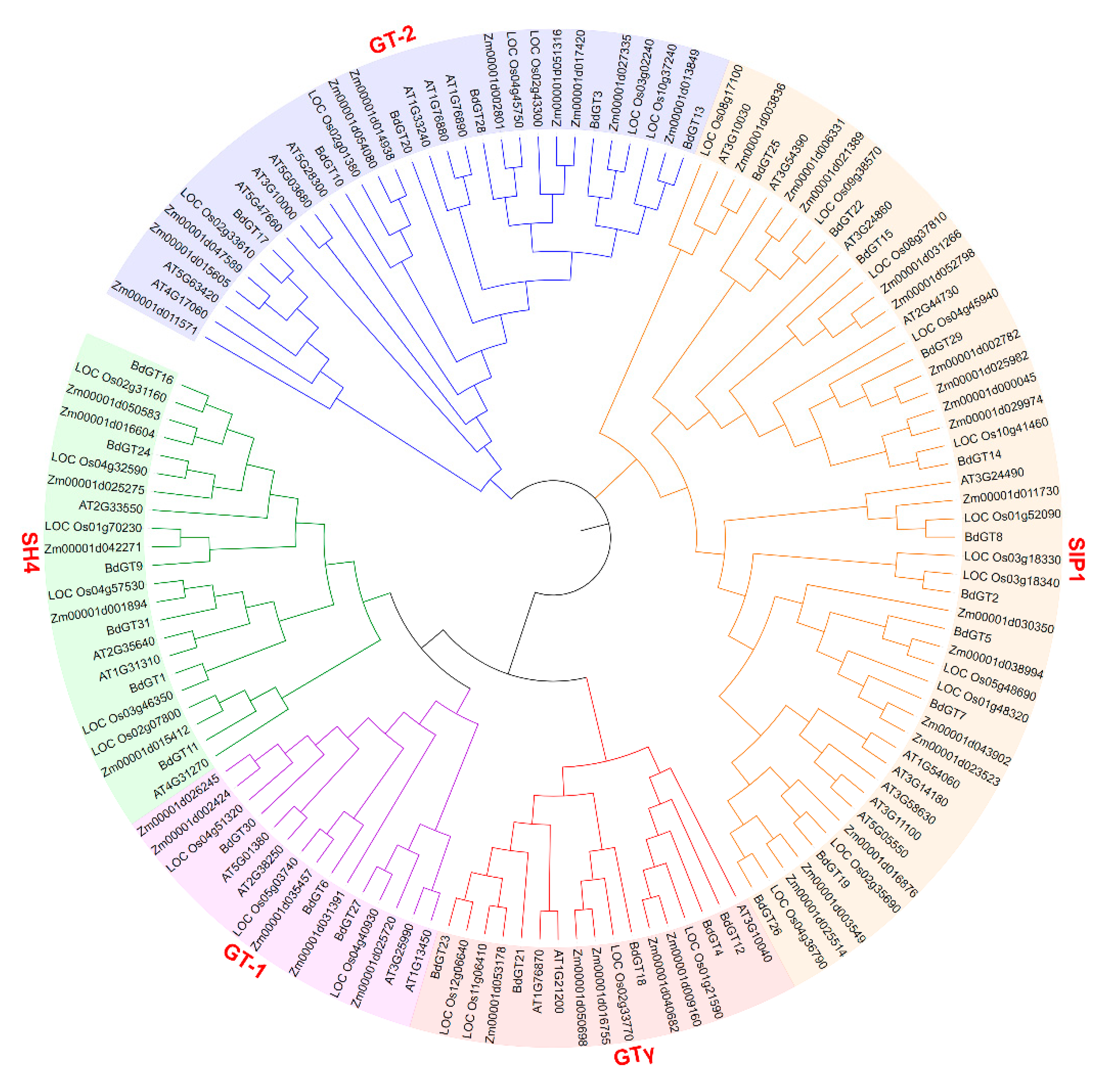
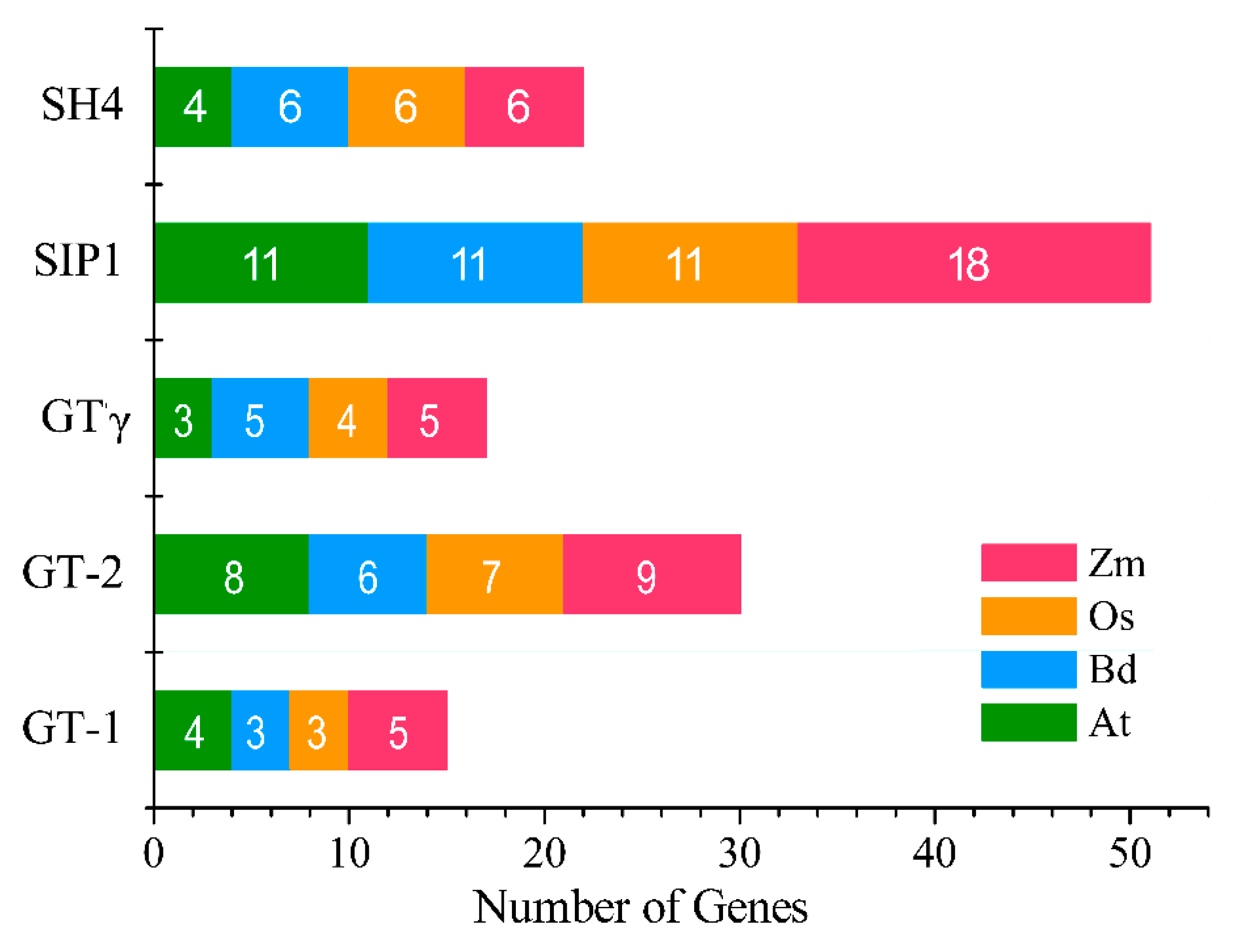
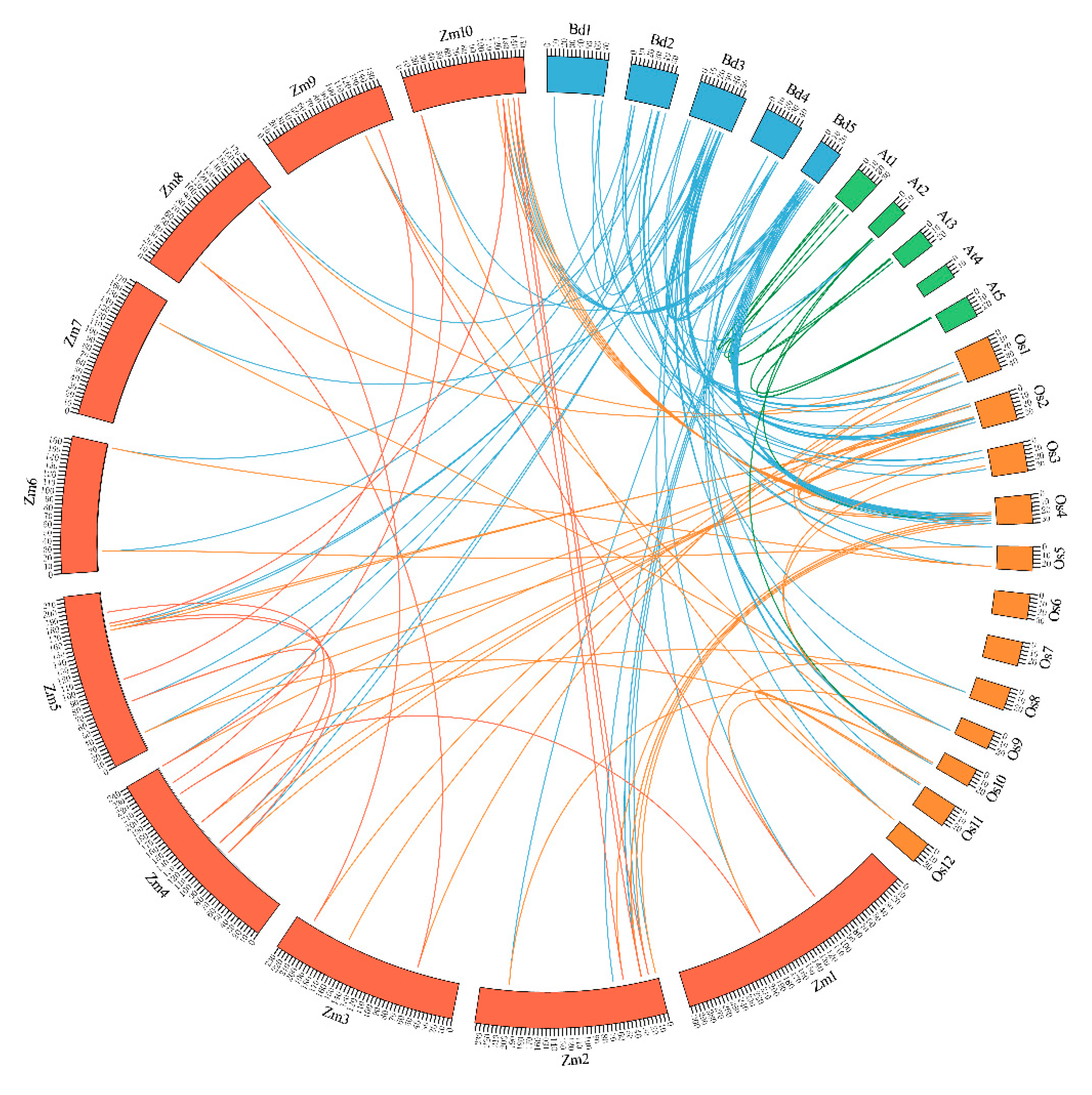
2.3. Phylogenetics and Synteny Analysis of BdGTs
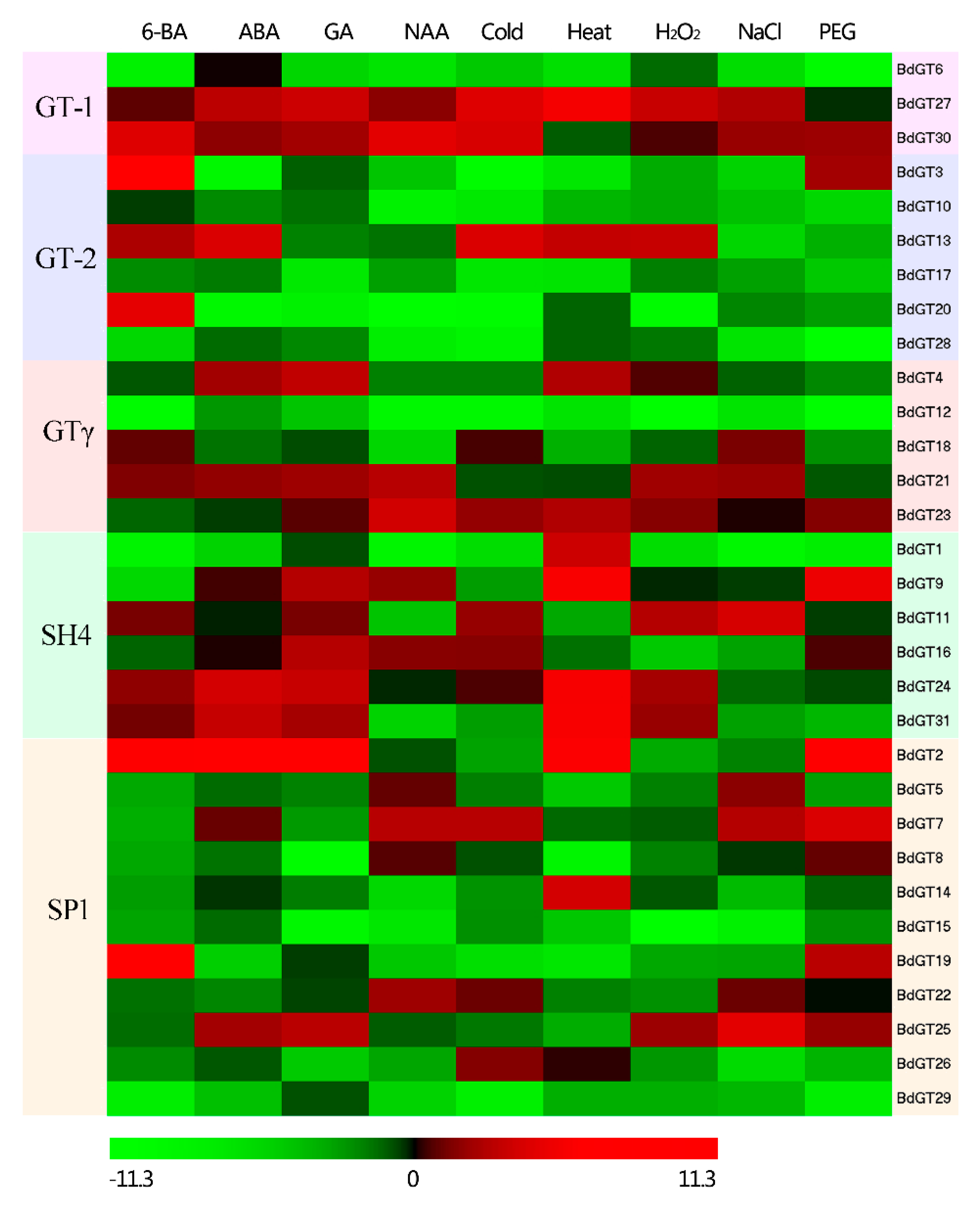
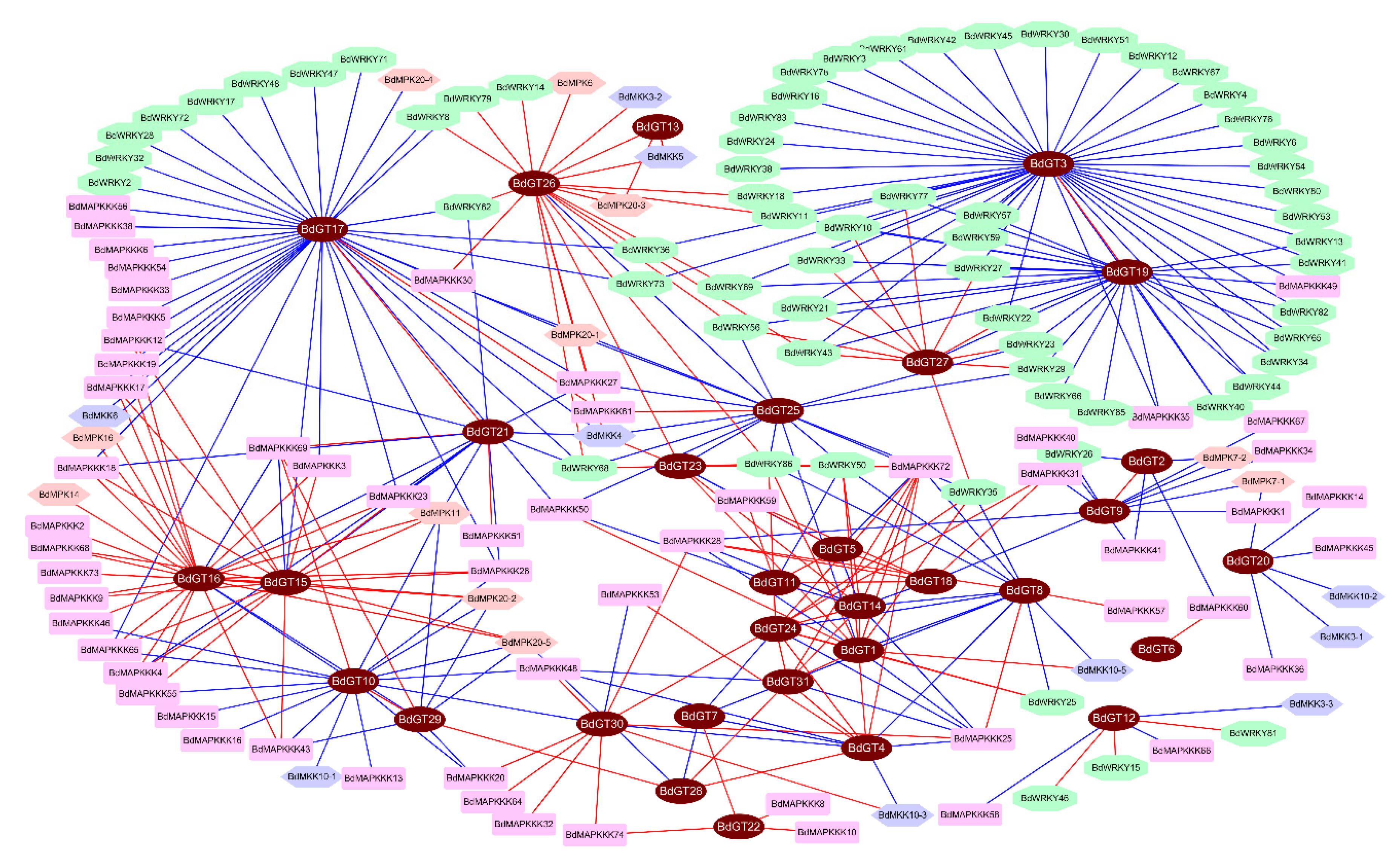
2.4. Expression Profile and Co-Regulatory Network of BdGT Genes in Response to Hormone and Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization Analysis of B. Distachyon GT Genes
4.2. Gene Structure and Chromosomal Locations
4.3. Protein Structure and Conserved Motif Analysis
4.4. Expression Analysis of BdGTs
4.5. Predicted Co-Expression Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TFs | Transcription factors |
| 6-BA | 6-benzylaminopurine |
| ABA | abscisic acid |
| NAA | 1-naphthylacetic acid |
| GA3 | gibberellin A3 |
| PEG | polyethylene glycol |
| PCCs | Pearson correlation coefficients |
References
- Hahn, M.W.; Pizer, M.; Wray, G.A.; Abouheif, E.; Balhoff, J.P.; Rockman, M.V.; Romano, L.A. The Evolution of Transcriptional Regulation in Eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Green, P.J.; Kay, S.A.; Chua, N.H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987, 6, 2543–2549. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.M.; Zou, H.F.; Lei, G.; Wei, W.; Zhou, Q.Y.; Niu, C.F.; Liao, Y.; Tian, A.G.; Ma, B.; Zhang, W.K.; et al. Soybean Trihelix Transcription Factors GmGT-2A and GmGT-2B Improve Plant Tolerance to Abiotic Stresses in Transgenic Arabidopsis. PLoS ONE 2009, 4, e6898. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. MGG 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Gao, M.J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of Seed Maturation Genes by a Trihelix Transcriptional Repressor in Arabidopsis Seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef]
- Osorio, M.B.; Bücker-Neto, L.; Castilhos, G.; Turchetto-Zolet, A.C.; Wiebke-Strohm, B.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 2012, 35, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; et al. Genome-Wide Characterization and Identification of Trihelix Transcription Factor and Expression Profiling in Response to Abiotic Stresses in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 251. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors. Int. J. Mol. Sci. 2016, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.L.; Zhao, N.; Lu, C.M. Plant Trihelix transcription factors family. Yi Chuan Hered. 2012, 34, 1551–1560. [Google Scholar] [CrossRef]
- Macrae, T.H.; Liu, L.; White, M.J. Transcription factors and their genes in higher plants. Functional domains, evolution and regulation. JBIC J. Biol. Inorg. Chem. 1999, 262, 247–257. [Google Scholar]
- Kuhn, R.M.; Caspar, T.; Dehesh, K.; Quail, P.H. DNA binding factor GT-2 from Arabidopsis. Plant Mol. Biol. 1993, 23, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y. Several Features of the GT-Factor Trihelix Domain Resemble Those of the Myb DNA-Binding Domain. Plant Physiol. 2000, 124, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Niyada, E.; Fujimoto, N.; Nagasaki, Y.; Noto, K.; Miyanoiri, Y.; Murata, J.; Hiratsuka, K.; Katahira, M. Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: Mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins Struct. Funct. Bioinform. 2010, 78, 3033–3047. [Google Scholar] [CrossRef]
- Kay, S.A.; Keith, B.; Shinozaki, K.; Chye, M.L.; Chua, N.H. The rice phytochrome gene: Structure, autoregulated expression, and binding of GT-1 to a conserved site in the 5’ upstream region. Plant Cell 1989, 1, 351–360. [Google Scholar]
- Perisic, O.; Lam, E. A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 1992, 4, 831–838. [Google Scholar]
- Le Gourrierec, J.; Li, Y.; Zhou, D. Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA-TBP-TATA complex. Plant J. 1999, 18, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Li, Q.T.; Chen, H.W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol. 2014, 14, 339. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.J. A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol. Biol. 2008, 66, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 Transcription Factor Regulates Water Use Efficiency and Drought Tolerance by Modulating Stomatal Density via Transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Tang, S.; Mei, F.; Peng, X.; Li, J.; Li, X.; Yan, X.; Zeng, X.; Liu, F.; Wu, Y.; et al. BnSIP1-1, a Trihelix Family Gene, Mediates Abiotic Stress Tolerance and ABA Signaling in Brassica napus. Front. Plant Sci. 2017, 8, 44. [Google Scholar] [CrossRef]
- Wang, R.; Hong, G.; Han, B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 2004, 324, 105–115. [Google Scholar] [CrossRef]
- Li, B.; Jiang, S.; Yu, X.; Cheng, C.; Chen, S.; Cheng, Y.; Yuan, J.S.; Jiang, D.; He, P.; Shan, L. Phosphorylation of Trihelix Transcriptional Repressor ASR3 by MAP KINASE4 Negatively Regulates Arabidopsis Immunity. Plant Cell 2015, 27, 839–856. [Google Scholar] [CrossRef]
- Völz, R.; Kim, S.K.; Mi, J.; Mariappan, K.G.; Guo, X.; Bigeard, J.; Alejandro, S.; Pflieger, D.; Rayapuram, N.; Al-Babili, S.; et al. The Trihelix transcription factor GT2-like 1 (GTL1) promotes salicylic acid metabolism, and regulates bacterial-triggered immunity. PLoS Genet. 2018, 14, e1007708. [Google Scholar] [CrossRef]
- Lv, D.W.; Li, X.; Zhang, M.; Gu, A.Q.; Zhen, S.M.; Wang, C.; Li, X.H.; Yan, Y.M. Large-scale phosphoproteome analysis in seedling leaves of Brachypodium distachyon L. BMC Genom. 2014, 15, 375. [Google Scholar] [CrossRef]
- Fan, M.; Herburger, K.; Jensen, J.K.; Zemelis-Durfee, S.; Brandizzi, F.; Fry, S.C.; Wilkerson, C.G. A Trihelix Family Transcription Factor Is Associated with Key Genes in Mixed-Linkage Glucan Accumulation. Plant Physiol. 2018, 178, 1207–1221. [Google Scholar] [CrossRef]
- Grimplet, J.; Deluc, L.G.; Tillett, R.L.; Wheatley, M.D.; Schlauch, K.A.; Cramer, G.R.; Cushman, J.C. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genom. 2007, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Wang, K.; Guo, T.; Jones, D.C.; Cobb, J.; Zhang, B.; Wang, Q. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genom. 2015, 15, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, F.; Cheng, L.; Kong, F.; Peng, Z.; Liu, S.; Yu, X.; Lu, G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S. Plant Productivity and Environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, s165–s183. [Google Scholar] [CrossRef]
- Gilmartin, P.M.; Memelink, J.; Hiratsuka, K.; Kay, S.A.; Chua, N.H. Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 1992, 4, 839–849. [Google Scholar]
- Quon, T.; O’Brien, M.; Sappl, P.G.; Smyth, D.R.; Kaplan-Levy, R.N.; Kaplan-Levy, R.N. Functional domains of the PETAL LOSS protein, a trihelix transcription factor that represses regional growth inArabidopsis thaliana. Plant J. 2014, 79, 477–491. [Google Scholar]
- Wang, Z.; Liu, Q.; Wang, H.; Zhang, H.; Xu, X.; Li, C.; Yang, C. Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci. Rep. 2016, 6, 36274. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, H.; Ji, H.; Wang, Y.; Dong, B.; Qiao, Y.; Liu, M.; Li, X. The Wheat GT Factor TaGT2L1D Negatively Regulates Drought Tolerance and Plant Development. Sci. Rep. 2016, 6, 27042. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Schröder, R.; Hallett, I.C.; Cohen, D.; Macrae, E.A. Overexpression of Polygalacturonase in Transgenic Apple Trees Leads to a Range of Novel Phenotypes Involving Changes in Cell Adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Fabi, J.P.; Cordenunsi, B.R.; Seymour, G.B.; Lajolo, F.M.; Nascimento, J.R.O.D. Molecular cloning and characterization of a ripening-induced polygalacturonase related to papaya fruit softening. Plant Physiol. Biochem. 2009, 47, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y.; Inaba, T.; Furuhashi, H.; Sasaki, Y. Trihelix DNA-binding Protein with Specificities for Two Distinctcis-Elements: Both important for light down-regulated and dark-inducible gene expression in higher plants. J. Biol. Chem. 2001, 276, 22238–22243. [Google Scholar] [CrossRef] [PubMed]
- Lam, E. Domain analysis of the plant DNA-binding protein GT1a: Requirement of four putative alpha-helices for DNA binding and identification of a novel oligomerization region. Mol. Cell. Biol. 1995, 15, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated Syntenic and Phylogenomic Analyses Reveal an Ancient Genome Duplication in Monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Qiu, Y.; Du, L.; Poovaiah, B. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 2012, 185, 274–280. [Google Scholar] [CrossRef]
- Chen, L.; Ren, F.; Zhong, H.; Feng, Y.; Jiang, W.; Li, X. Identification and expression analysis of genes in response to high-salinity and drought stresses in Brassica napus. Acta Biochim. Biophys. Sin. 2010, 42, 154–164. [Google Scholar] [CrossRef]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-Induced Expression of the SCaM-4 Promoter Is Mediated in Part by a GT-1 Box That Interacts with a GT-1-Like Transcription Factor1. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Viana, V.E.; Busanello, C.; Da Maia, L.C.; Pegoraro, C.; De Oliveira, A.C. Activation of rice WRKY transcription factors: An army of stress fighting soldiers? Curr. Opin. Plant Biol. 2018, 45, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ye, N.; Cao, J.; Jiang, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BiorXiv 2018, 289660. [Google Scholar] [CrossRef]
- Sun, H.; Wei, H.; Wang, H.; Hao, P.; Gu, L.; Liu, G.; Ma, L.; Su, Z.; Yu, S. Genome-wide identification and expression analysis of the BURP domain-containing genes in Gossypium hirsutum. BMC Genom. 2019, 20, 558. [Google Scholar] [CrossRef]
- Liu, Z.; Ge, X.; Yang, Z.; Zhang, C.; Zhao, G.; Chen, E.; Liu, J.; Zhang, X.; Li, F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 2017, 18, 54. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, X.; Li, T.; Jia, M.; Liu, X.; Zou, Y.; Liu, Z.; Wen, F. Identification, characterization, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon. Funct. Integr. Genom. 2018, 18, 709–724. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.Y.; He, J.E.; Lanczycki, C.J.; Lu, S.N.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Zhu, H.; Li, P.; Jiang, M.; Mao, W.; Ong, C.; Chu, Z. Genome-Wide Evolutionary Characterization and Expression Analyses of WRKY Family Genes in Brachypodium distachyon. DNA Res. 2014, 21, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene Locus | Chr | ORF (bp) | Exon No. | PI | MW (kD) | Length (aa) | Subcellular Localization | GT Domain |
|---|---|---|---|---|---|---|---|---|---|
| BdGT1 | Bradi1g12900 | Chr1 | 1026 | 2 | 8.68 | 37.20 | 341 | nucl | 44–146 |
| BdGT2 | Bradi1g65400 | Chr1 | 780 | 1 | 9.65 | 27.84 | 259 | chlo/mito | 19–112 |
| BdGT3 | Bradi1g77610 | Chr1 | 2130 | 3 | 8.75 | 75.46 | 709 | nucl | 76–162, 419–515 |
| BdGT4 | Bradi2g12030 | Chr2 | 1662 | 1 | 6.28 | 59.29 | 553 | nucl | 109–262 |
| BdGT5 | Bradi2g16780 | Chr2 | 1068 | 1 | 9.92 | 37.91 | 355 | nucl | 41–130 |
| BdGT6 | Bradi2g38230 | Chr2 | 1038 | 1 | 6.07 | 38.46 | 345 | nucl | 50–163 |
| BdGT7 | Bradi2g46320 | Chr2 | 963 | 1 | 9.8 | 35.03 | 320 | nucl | 17–106 |
| BdGT8 | Bradi2g48320 | Chr2 | 912 | 1 | 6.57 | 33.82 | 303 | nucl | 57–153 |
| BdGT9 | Bradi2g59440 | Chr2 | 948 | 3 | 5.75 | 34.50 | 315 | chlo/nucl | 12–145 |
| BdGT10 | Bradi3g00697 | Chr3 | 2277 | 2 | 5.88 | 80.53 | 758 | nucl | 238–325, 555–643 |
| BdGT11 | Bradi3g05530 | Chr3 | 1104 | 2 | 4.23 | 38.37 | 367 | nucl/cyto | 80–189 |
| BdGT12 | Bradi3g17539 | Chr3 | 1452 | 1 | 7.62 | 52.38 | 483 | cyto | 102–251 |
| BdGT13 | Bradi3g30457 | Chr3 | 2310 | 3 | 5.72 | 82.09 | 769 | nucl | 87–173, 454–541 |
| BdGT14 | Bradi3g33630 | Chr3 | 975 | 1 | 9.53 | 34.30 | 324 | nucl/chlo | 60–152 |
| BdGT15 | Bradi3g38682 | Chr3 | 1098 | 2 | 6.25 | 40.60 | 365 | nucl | 35–125 |
| BdGT16 | Bradi3g44370 | Chr3 | 1056 | 2 | 5.12 | 37.76 | 351 | chlo/nucl | 26–148 |
| BdGT17 | Bradi3g45230 | Chr3 | 2628 | 17 | 8.9 | 96.63 | 875 | chlo | 116–216, 782–865 |
| BdGT18 | Bradi3g45300 | Chr3 | 1203 | 1 | 5.77 | 46.18 | 400 | nucl | 74–205 |
| BdGT19 | Bradi3g46210 | Chr3 | 1212 | 2 | 6.54 | 43.22 | 403 | nucl | 62–147 |
| BdGT20 | Bradi3g50213 | Chr3 | 4176 | 20 | 4.47 | 152.26 | 1391 | chlo | 82–168, 377–454 |
| BdGT21 | Bradi4g24750 | Chr4 | 1269 | 1 | 7.01 | 47.83 | 422 | nucl | 103–234 |
| BdGT22 | Bradi4g37730 | Chr4 | 1035 | 1 | 7.56 | 36.98 | 344 | nucl | 29–135 |
| BdGT23 | Bradi4g41830 | Chr4 | 1290 | 1 | 5.76 | 49.11 | 429 | nucl | 98–229 |
| BdGT24 | Bradi5g08600 | Chr5 | 981 | 3 | 6.3 | 35.24 | 326 | nucl/cyto | 17–134 |
| BdGT25 | Bradi5g08980 | Chr5 | 1509 | 7 | 7.53 | 53.76 | 502 | nucl | 114–206 |
| BdGT26 | Bradi5g11070 | Chr5 | 1257 | 1 | 6.97 | 45.34 | 418 | nucl | 75–160 |
| BdGT27 | Bradi5g13900 | Chr5 | 1140 | 5 | 5.96 | 41.40 | 379 | chlo/nucl | 48–138 |
| BdGT28 | Bradi5g17150 | Chr5 | 2277 | 3 | 6.11 | 81.89 | 758 | nucl/pero | 86–172, 497–584 |
| BdGT29 | Bradi5g17281 | Chr5 | 597 | 1 | 11.07 | 21.82 | 198 | chlo/nucl | 23–98 |
| BdGT30 | Bradi5g20847 | Chr5 | 828 | 2 | 8.97 | 32.09 | 275 | nucl | 17–107 |
| BdGT31 | Bradi5g25700 | Chr5 | 1236 | 2 | 10.05 | 44.26 | 411 | chlo/nucl | 78–215 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, F.; Xu, L.; Xie, Y.; Liao, L.; Li, T.; Jia, M.; Liu, X.; Wu, X. Identification and in Silico Characterization of GT Factors Involved in Phytohormone and Abiotic Stresses Responses in Brachypodium distachyon. Int. J. Mol. Sci. 2019, 20, 4115. https://doi.org/10.3390/ijms20174115
Wen F, Xu L, Xie Y, Liao L, Li T, Jia M, Liu X, Wu X. Identification and in Silico Characterization of GT Factors Involved in Phytohormone and Abiotic Stresses Responses in Brachypodium distachyon. International Journal of Molecular Sciences. 2019; 20(17):4115. https://doi.org/10.3390/ijms20174115
Chicago/Turabian StyleWen, Feng, Liangwei Xu, Yuebin Xie, Liang Liao, Tongjian Li, Mingliang Jia, Xinsheng Liu, and Xiaozhu Wu. 2019. "Identification and in Silico Characterization of GT Factors Involved in Phytohormone and Abiotic Stresses Responses in Brachypodium distachyon" International Journal of Molecular Sciences 20, no. 17: 4115. https://doi.org/10.3390/ijms20174115
APA StyleWen, F., Xu, L., Xie, Y., Liao, L., Li, T., Jia, M., Liu, X., & Wu, X. (2019). Identification and in Silico Characterization of GT Factors Involved in Phytohormone and Abiotic Stresses Responses in Brachypodium distachyon. International Journal of Molecular Sciences, 20(17), 4115. https://doi.org/10.3390/ijms20174115





