Cellular Metabolomics Reveal the Mechanism Underlying the Anti-Atherosclerotic Effects of Aspirin Eugenol Ester on Vascular Endothelial Dysfunction
Abstract
:1. Introduction
2. Results
2.1. AEE Reduced the H2O2-Induced Apoptosis of HUVECs
2.2. AEE Ameliorated Alterations in the Biochemical Profile
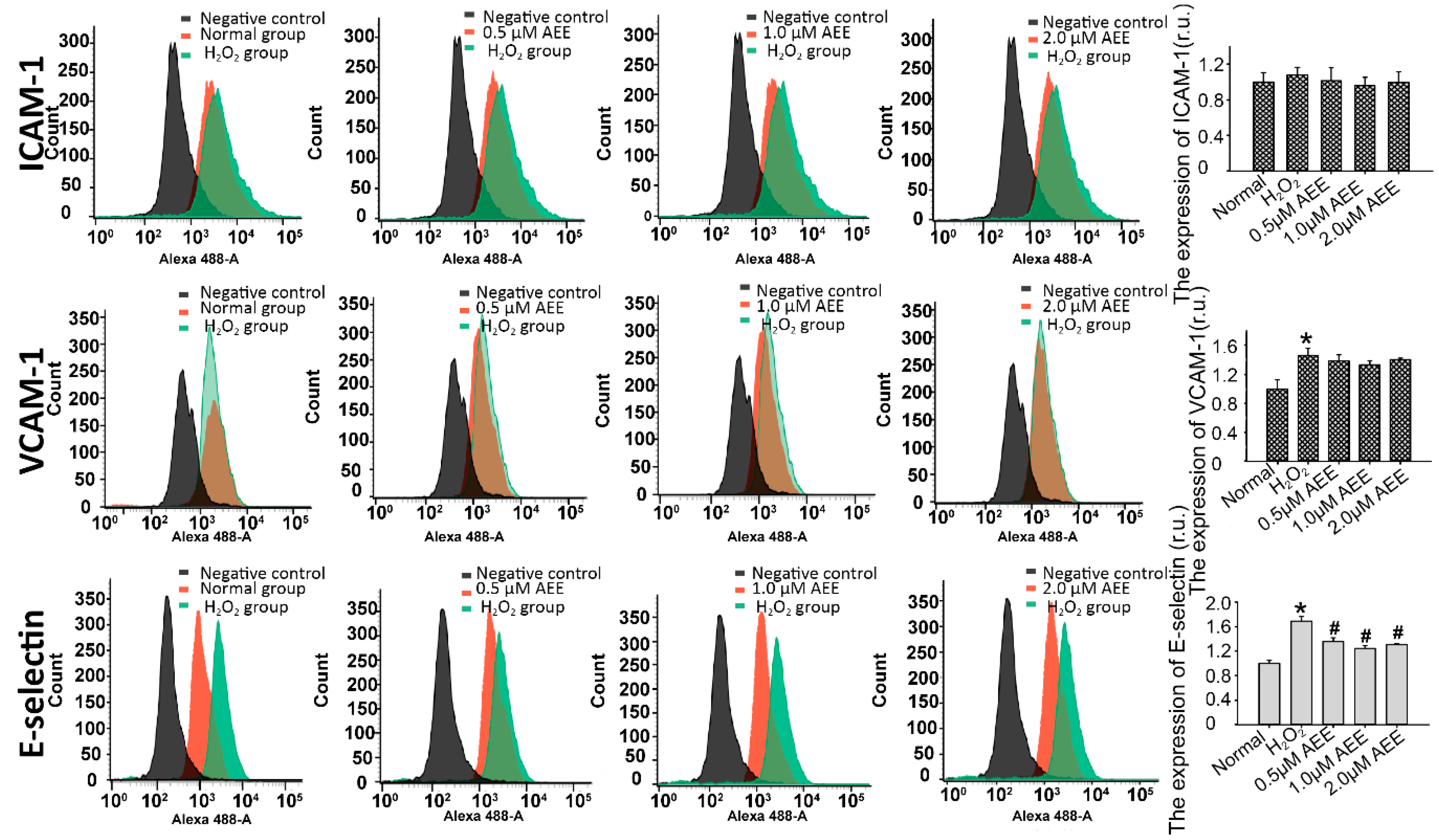
2.3. AEE Suppressed the Oxidative Injury-Induced Imbalance in Vascular Adhesion
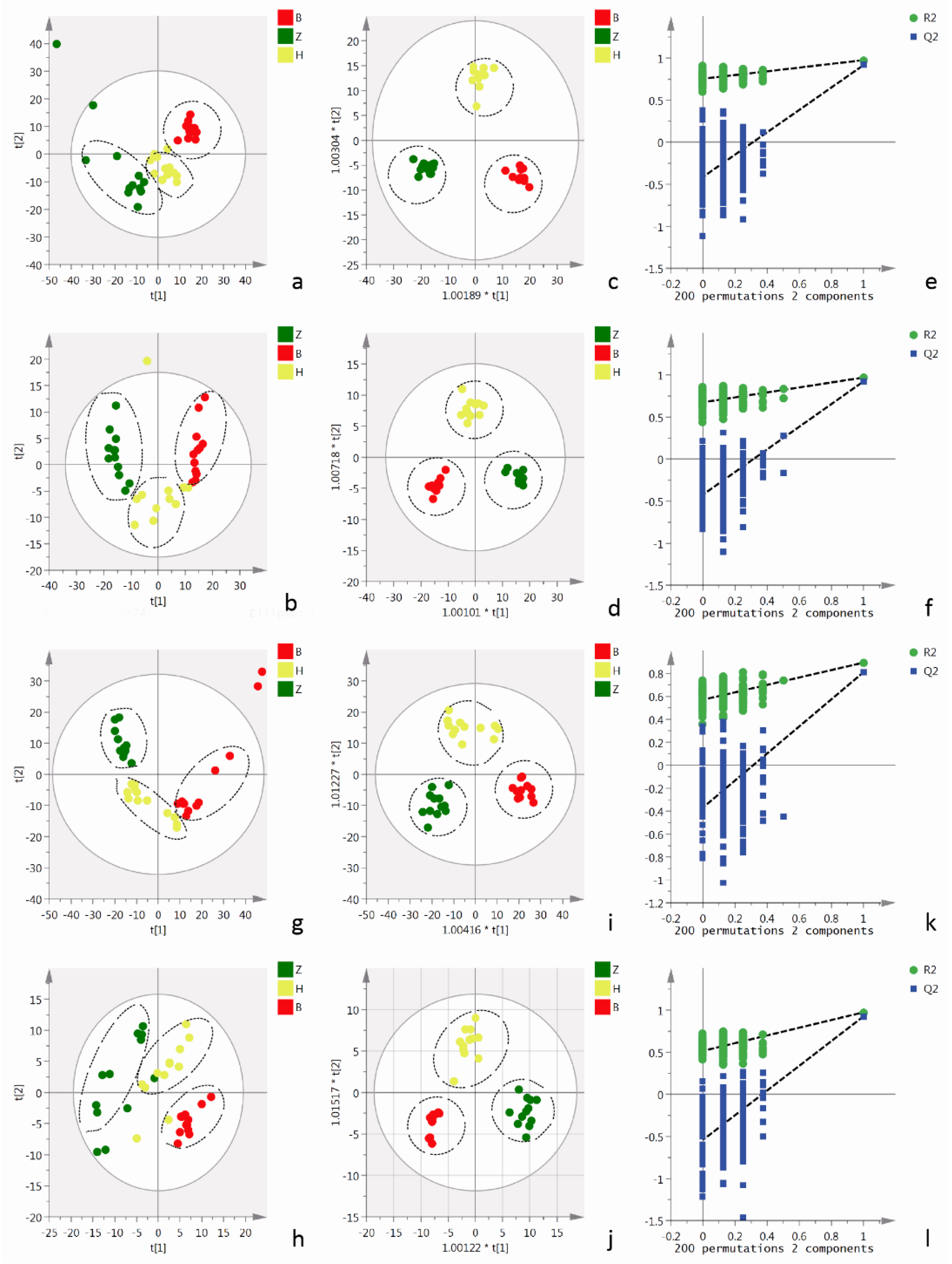
2.4. Metabolomics Analysis of Cells and Cell Culture Supernatants
2.4.1. Metabolomics Analysis of Cell Culture Supernatants
2.4.2. Cellular Metabolomics Analysis
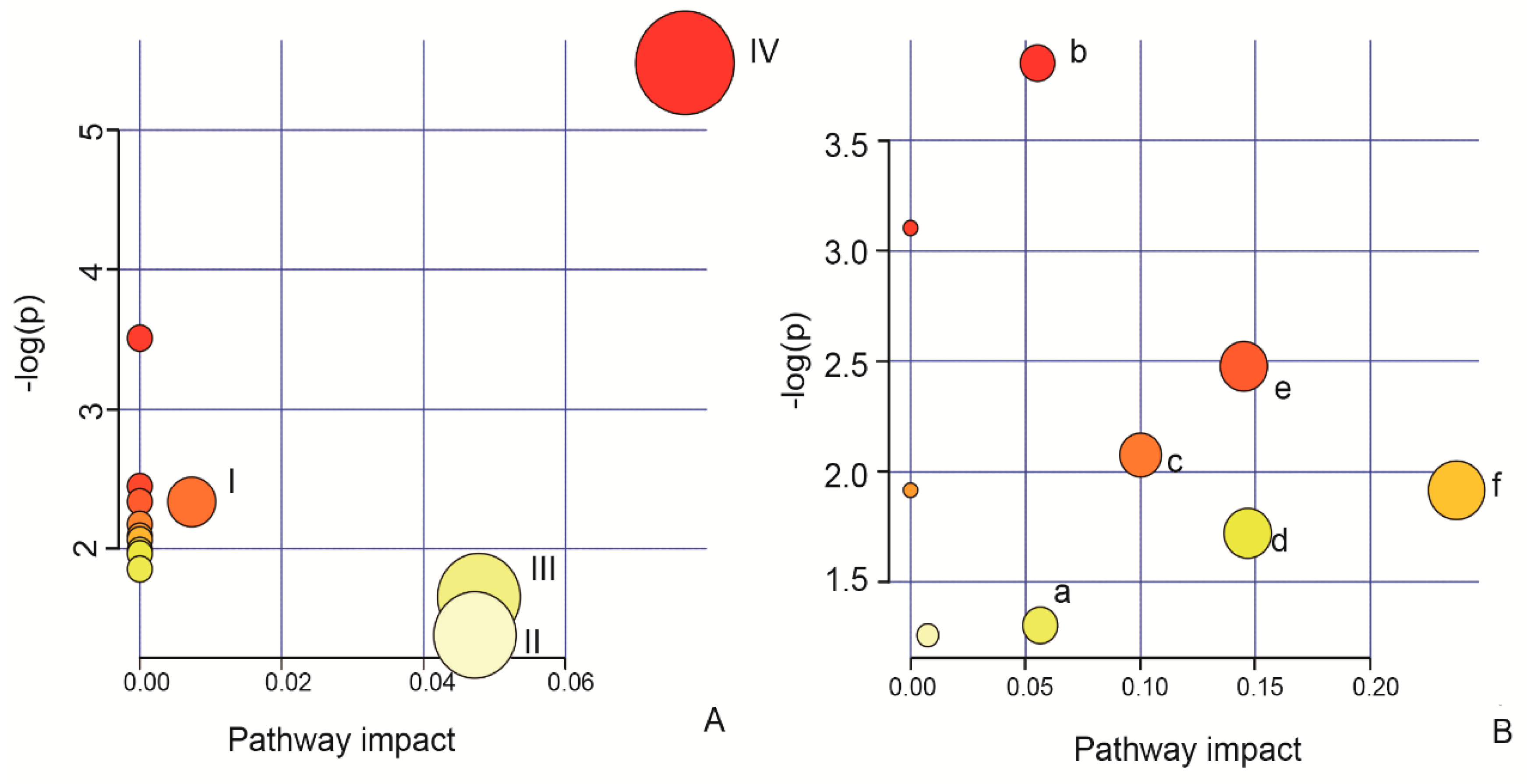
2.5. Identification of the Biomarkers and Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Treatments
4.3. Apoptosis Detection Using Flow Cytometry
4.4. Adhesion Assay
4.5. Adhesion Molecule Expression
4.6. Cellular Metabolite Extraction
4.7. UPLC/QTOF-MS Analysis of Cellular Metabolites
4.8. Animal Experiment
4.9. Measurement of Serum AGE Levels
4.10. Adhesion Molecule Expression in the Aorta
4.11. Serum Levels of Adhesion Molecules
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HUVECs | Human umbilical vein endothelial cells |
| HFD | High fat diet |
| VCAM-1 | Vascular cell adhesion molecule 1 |
| ICAM-1 | Soluble intercellular adhesion molecule 1 |
| MDA | Malondialdehyde |
| HDL | High density lipoprotein |
| AGEs | Advanced glycosylation end products |
| LDL | Low density lipoprotein |
| TCH | Total cholesterol |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal projections to latent structures discriminate analysis |
| VIP | Variable importance in the projection |
| UPLC/QTOF-MS | Ultra-performance liquid chromatography/quadrupole time of flight-mass spectrometry |
References
- Henderson, J.; Henderson, I.W. The endocrine function of the vascular endothelium. J. Biol. Educ. 1995, 29, 104–109. [Google Scholar] [CrossRef]
- Hurairah, H.; Ferro, A. The role of the endothelium in the control of vascular function. Int. J. Clin. Pract. 2004, 58, 173–183. [Google Scholar] [CrossRef]
- Henrion, D. Endoplasmic reticulum stress and vascular endothelium dysfunction. Fundam. Clin. Pharmacol. 2013, 27, 4. [Google Scholar]
- Jamwal, S.; Sharma, S. Vascular endothelium dysfunction: A conservative target in metabolic disorders. Inflamm. Res. 2018, 67, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Markov, C.M. Molecular mechanisms of dysfunction of vascular endothelium. Kardiologiya 2005, 45, 62–72. [Google Scholar]
- Vestweber, D. How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 2015, 15, 692–704. [Google Scholar] [CrossRef]
- Cai, J.J.; Wen, J.; Jiang, W.H.; Lin, J.; Hong, Y.; Zhu, Y.S. Androgen actions on endothelium functions and cardiovascular diseases. J. Geriatr. Cardiol. 2016, 13, 183–196. [Google Scholar]
- Lin, P.J.; Chang, C.H. Endothelium dysfunction in cardiovascular diseases. Chang Yi Xue Za Zhi 1994, 17, 198–210. [Google Scholar]
- Nair, A.R.; Agbor, L.N.; Mukohda, M.; Hu, C.Y.; Wu, J.; Sigmund, C.D. Interference with ppar gamma in the endothelium causes cerebral vascular dysfunction in response to endogenous renin-angiotensin system activation. FASEB J. 2017, 31. [Google Scholar]
- Onyangunga, O.; Moodley, J.; Odun-Ayo, F.; Naicker, T. The impact of antiretroviral drugs in placental vascular endothelium dysfunction. Placenta 2017, 57, 273–274. [Google Scholar] [CrossRef]
- Li, J.Y.; Yu, Y.G.; Wang, Q.W.; Zhang, J.Y.; Yang, Y.J.; Li, B.; Zhou, X.Z.; Niu, J.R.; Wei, X.J.; Liu, X.W.; et al. Synthesis of aspirin eugenol ester and its biological activity. Med. Chem. Res. 2012, 21, 995–999. [Google Scholar] [CrossRef]
- Ma, N.; Liu, X.W.; Yang, Y.J.; Li, J.Y.; Mohamed, I.; Liu, G.R.; Zhang, J.Y. Preventive effect of aspirin eugenol ester on thrombosis in kappa-carrageenan-induced rat tail thrombosis model. PLoS ONE 2015, 10, e0133125. [Google Scholar]
- Ma, N.; Liu, X.W.; Yang, Y.J.; Shen, D.S.; Zhao, X.L.; Mohamed, I.; Kong, X.J.; Li, J.Y. Evaluation on antithrombotic effect of aspirin eugenol ester from the view of platelet aggregation, hemorheology, txb2/6-keto-pgf1alpha and blood biochemistry in rat model. BMC Vet. Res. 2016, 12, 108. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Yang, G.Z.; Liu, X.W.; Yang, Y.J.; Mohamed, I.; Liu, G.R.; Li, J.Y. Impact of aspirin eugenol ester on cyclooxygenase-1, cyclooxygenase-2, c-reactive protein, prothrombin and arachidonate 5-lipoxygenase in healthy rats. Iran. J. Pharm. Res. 2017, 16, 1443–1451. [Google Scholar] [PubMed]
- Ma, N.; Yang, Y.; Liu, X.; Kong, X.; Li, S.; Qin, Z.; Jiao, Z.; Li, J. Uplc-q-tof/ms-based metabonomic studies on the intervention effects of aspirin eugenol ester in atherosclerosis hamsters. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Li, J.; Yu, Y.; Yang, Y.; Liu, X.; Zhang, J.; Li, B.; Zhou, X.; Niu, J.; Wei, X. Antioxidant activity of aspirin eugenol ester for aging model of mice by d-galactose. J. Anim. Vet. Adv. 2012, 11, 4401–4405. [Google Scholar]
- Ma, N.; Karam, I.; Liu, X.W.; Kong, X.J.; Qin, Z.; Li, S.H.; Jiao, Z.H.; Dong, P.C.; Yang, Y.J.; Li, J.Y. Uplc-q-tof/ms-based urine and plasma metabonomics study on the ameliorative effects of aspirin eugenol ester in hyperlipidemia rats. Toxicol. Appl. Pharmacol. 2017, 332, 40–51. [Google Scholar] [CrossRef]
- Ma, N.; Liu, X.W.; Kong, X.J.; Li, S.H.; Jiao, Z.H.; Qin, Z.; Yang, Y.J.; Li, J.Y. Aspirin eugenol ester regulates cecal contents metabolomic profile and microbiota in an animal model of hyperlipidemia. BMC Vet. Res. 2018, 14, 405. [Google Scholar] [CrossRef]
- Baharum, S.N.; Azizan, K.A. Metabolomics in systems biology. Adv. Exp. Med. Biol. 2018, 1102, 51–68. [Google Scholar]
- Balashova, E.E.; Maslov, D.L.; Lokhov, P.G. A metabolomics approach to pharmacotherapy personalization. J. Pers. Med. 2018, 8. [Google Scholar] [CrossRef]
- Kargul, J.; Irminger-Finger, I.; Laurent, G.J. Metabolomics: Taking snapshots of cellular physiology in health and disease. Int. J. Biochem. Cell Biol. 2017, 93, 86. [Google Scholar] [CrossRef] [PubMed]
- Lains, I.; Gantner, M.; Murinello, S.; Lasky-Su, J.A.; Miller, J.W.; Friedlander, M.; Husain, D. Metabolomics in the study of retinal health and disease. Prog. Retin. Eye Res. 2019, 69, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Olesti, E.; De Toma, I.; Ramaekers, J.G.; Brunt, T.M.; Carbo, M.L.; Fernandez-Aviles, C.; Robledo, P.; Farre, M.; Dierssen, M.; Pozo, O.J.; et al. Metabolomics predicts the pharmacological profile of new psychoactive substances. J. Psychopharmacol. 2019, 33, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wei, J.; Liu, Y.; Pei, D.; Hu, Q.; Wang, Y.; Di, D. Discovery of biomarkers for oxidative stress based on cellular metabolomics. Biomarkers 2016, 21, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Ohashi, Y.; Soga, T.; Tomita, M. Unveiling cellular biochemical reactions via metabolomics-driven approaches. Curr. Opin. Microbiol. 2010, 13, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Silveri, F.; De Angelis, R.; Argentati, F.; Brecciaroli, D.; Muti, S.; Cervini, C. Hypertrophic osteoarthropathy: Endothelium and platelet function. Clin. Rheumatol. 1996, 15, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.M.; Kaiser, E.; Nawroth, P.P. Regulation of the coagulation system by vascular endothelial cells. Haemostasis 1988, 18, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Butta, N.V.; Fernandez-Bello, I.; Lopez-Longo, F.J.; Jimenez-Yuste, V. Endothelial dysfunction and altered coagulation as mediators of thromboembolism in behcet disease. Semin. Thromb. Hemost. 2015, 41, 621–628. [Google Scholar] [PubMed]
- Ivanciu, L.; Stalker, T.J. Spatiotemporal regulation of coagulation and platelet activation during the hemostatic response in vivo. J. Thromb. Haemost. 2015, 13, 1949–1959. [Google Scholar] [CrossRef]
- Mensah, G.A.; Ryan, U.S.; Hooper, W.C.; Engelgau, M.M.; Callow, A.D.; Kapuku, G.K.; Mantovani, A. Vascular endothelium summary statement ii: Cardiovascular disease prevention and control. Vasc. Pharm. 2007, 46, 318–320. [Google Scholar] [CrossRef]
- Ramli, J.; CalderonArtero, P.; Block, R.C.; Mousa, S.A. Novel therapeutic targets for preserving a healthy endothelium: Strategies for reducing the risk of vascular and cardiovascular disease. Cardiol. J. 2011, 18, 352–363. [Google Scholar] [PubMed]
- Heitzer, T.; Schlinzig, T.; Krohn, K.; Meinertz, T.; Munzel, T. Endothelial dysfunction, oxidative stress, and risk of cardiovascular events in patients with coronary artery disease. Circulation 2001, 104, 2673–2678. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.; Kuehn, S.; Jashari, A.; Tsai, T.; Bartz-Schmidt, K.U.; Schnichels, S.; Joachim, S.C. A novel porcine ex vivo retina culture model for oxidative stress induced by h(2)o(2). Altern. Lab. Anim. 2017, 45, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Kaczara, P.; Sarna, T.; Burke, J.M. Dynamics of h2o2 availability to arpe-19 cultures in models of oxidative stress. Free Radic. Biol. Med. 2010, 48, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Tang, M. Exercise improves high fat diet-impaired vascular function. Biomed. Rep. 2017, 7, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaprinay, B.; Liptak, B.; Slovak, L.; Svik, K.; Knezl, V.; Sotnikova, R.; Gasparova, Z. Hypertriglyceridemic rats fed high fat diet as a model of metabolic syndrome. Physiol. Res. 2016, 65, S515–S518. [Google Scholar]
- Karam, I.; Ma, N.; Liu, X.W.; Li, S.H.; Kong, X.J.; Li, J.Y.; Yang, Y.J. Regulation effect of aspirin eugenol ester on blood lipids in wistar rats with hyperlipidemia. BMC Vet. Res. 2015, 11. [Google Scholar] [CrossRef]
- Segeda, I.; Shevchuk, S.; Kuvikova, I. Soluble vascular cell adhesion molecule-1 level in patients with anthiphospolipid syndrome: Its association with disease activity and endothelium dysfunction. Rheumatology 2012, 51, 181. [Google Scholar]
- Chen, N.G.; Han, X. Dual function of troglitazone in icam-1 gene expression in human vascular endothelium. Biochem. Biophys. Res. Commun. 2001, 282, 717–722. [Google Scholar] [CrossRef]
- Hallahan, D.; Kuchibhotla, J.; Wyble, C. Cell adhesion molecules mediate radiation-induced leukocyte adhesion to the vascular endothelium. Cancer Res. 1996, 56, 5150–5155. [Google Scholar]
- Chi, Z.; Melendez, A.J. Role of cell adhesion molecules and immune-cell migration in the initiation, onset and development of atherosclerosis. Cell Adhes. Migr. 2007, 1, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Munshi, R.; Clanachan, A.S.; Baer, H.P. 5’-deoxy-5’-methylthioadenosine: A nucleoside which differentiates between adenosine receptor types. Biochem. Pharm. 1988, 37, 2085–2089. [Google Scholar] [CrossRef]
- Lazzaro, V.A.; Walker, R.J.; Duggin, G.G.; Phippard, A.; Horvath, J.S.; Tiller, D.J. Inhibition of fibroblast proliferation in l-valine reduced selective media. Res. Commun. Chem. Pathol. Pharmacol. 1992, 75, 39–48. [Google Scholar] [PubMed]
- Selamnia, M.; Robert, V.; Mayeur, C.; Duee, P.H.; Blachier, F. Effects of l-valine on growth and polyamine metabolism in human colon carcinoma cells. Biochim. Biophys Acta 1998, 1379, 151–160. [Google Scholar] [CrossRef]
- Bouhlal, H.; Blondin, C.; Haeffner-Cavaillon, N.; Goldschmidt, P. n acetyl-aspartyl glutamic acid (naaga) inhibits the adhesion of leukocytes to activated endothelial cells and down-modulates the cytokine-induced expression of adhesion molecules. J. Fr. D’ophtalmologie 2002, 25, 993–1000. [Google Scholar]
- Chen, J.; Wu, F.; Long, Y.; Yu, W. Glutathione supplementation attenuates oxidative stress and improves vascular hyporesponsiveness in experimental obstructive jaundice. Oxid. Med. Cell Longev. 2015, 2015, 486148. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, W.; Zhang, Y.; Wang, W.; Yao, F.; Yan, J.; Wan, C. 5’-methylthioadenosine attenuates ischemia reperfusion injury after liver transplantation in rats. Inflammation 2014, 37, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Simile, M.M.; Banni, S.; Angioni, E.; Carta, G.; De Miglio, M.R.; Muroni, M.R.; Calvisi, D.F.; Carru, A.; Pascale, R.M.; Feo, F. 5’-methylthioadenosine administration prevents lipid peroxidation and fibrogenesis induced in rat liver by carbon-tetrachloride intoxication. J. Hepatol. 2001, 34, 386–394. [Google Scholar] [CrossRef]
- Ashoori, M.; Saedisomeolia, A. Riboflavin (vitamin b(2)) and oxidative stress: A review. Br. J. Nutr. 2014, 111, 1985–1991. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zheng, Y.; Zhu, X.; Li, S.; Zhou, C. L-lysine and l-arginine inhibit the oxidation of lipids and proteins of emulsion sausage by chelating iron ion and scavenging radical. Asian-Australas. J. Anim. Sci. 2018, 31, 905–913. [Google Scholar] [CrossRef]
- Dou, L.; Bertrand, E.; Cerini, C.; Faure, V.; Sampol, J.; Vanholder, R.; Berland, Y.; Brunet, P. The uremic solutes p-cresol and indoxyl sulfate inhibit endothelial proliferation and wound repair. Kidney Int. 2004, 65, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, T.; Takahashi, K.; Kobayashi, T.; Oshima, E.; Iwasaki, S.; Suzuki, H. Oxidized low density lipoprotein (ox-ldl) as a marker of atherosclerosis in hemodialysis (hd) patients. Clin. Nephrol. 2002, 58, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Alique, M.; Luna, C.; Carracedo, J.; Ramirez, R. LDL biochemical modifications: A link between atherosclerosis and aging. Food Nutr. Res. 2015, 59. [Google Scholar] [CrossRef] [PubMed]
- Bartels, E.D.; Christoffersen, C.; Lindholm, M.W.; Nielsen, L.B. Altered metabolism of ldl in the arterial wall precedes atherosclerosis regression. Circ. Res. 2015, 117, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Schleicher, E.; Friess, U. Oxidative stress, age, and atherosclerosis. Kidney Int. 2007, 72, S17–S26. [Google Scholar] [CrossRef] [PubMed]
- McNair, E.; Qureshi, M.; Prasad, K.; Pearce, C. Atherosclerosis and the hypercholesterolemic age-rage axis. Int. J. Angiol. 2016, 25, 110–116. [Google Scholar]
- Yamagishi, S.I.; Matsui, T. Role of hyperglycemia-induced advanced glycation end product (age) accumulation in atherosclerosis. Ann. Vasc. Dis. 2018, 11, 253–258. [Google Scholar] [CrossRef]
- Xia, L.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids. Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Karam, I.; Ma, N.; Liu, X.W.; Kong, X.J.; Zhao, X.L.; Yang, Y.J.; Li, J.Y. Lowering effects of aspirin eugenol ester on blood lipids in rats with high fat diet. Lipids Health Dis. 2016, 15, 1–5. [Google Scholar] [CrossRef] [Green Version]



| Variables | Normal Group | HFD Group | AEE Group |
|---|---|---|---|
| HDL (mmol/L) | 0.96 ± 0.10 | 0.67 ± 0.06 * | 0.79 ± 0.02 # |
| LDL (mmol/L) | 0.53 ± 0.06 | 0.80 ± 0.19 * | 0.68 ± 0.04 # |
| TCH (mmol/L) | 1.24 ± 0.10 | 2.41 ± 0.48 * | 1.58 ± 0.34 # |
| AGE (ng/mL) | 390.38 ± 18.13 | 786.64 ± 49.20 * | 497.04 ± 34.70 # |
| No. | RT | VIP | Formula | Metabolite | SM | m/z | Fold Change |
|---|---|---|---|---|---|---|---|
| AEE/C | |||||||
| 1 | 3.14 | 3.39 | C9H11NO3 | l-Tyrosine | − | 181.07285 | 2.16 * |
| 2 | 1.49 | 1.01 | C5H11NO2 | l-valine | + | 117.07925 | 3.17 * |
| 3 | 10.56 | 2.19 | C7H12O5 | 2-Isopropylmalic acid | + | 176.06818 | 0.49 * |
| 4 | 5.7 | 4.093 | C11H15N5O3S | 5’-Methylthioadenosine | + | 297.08875 | 1.75 * |
| No. | RT | VIP | Formula | Metabolite | SM | m/z | Fold Change | |
|---|---|---|---|---|---|---|---|---|
| C/H2O2 | AEE/H2O2 | |||||||
| 1 | 1.46 | 1.45 | C10H17N3O6S | Glutathione | + − | 307.08353 | 2.78 * | 1.40 * |
| 2 | 1.14 | 1.27 | C6H14N2O2 | l-Lysine | + | 146.10437 | 6.17 * | 5.16 * |
| 3 | 2.04 | 4.13 | C11H16N2O8 | N-Acetylaspartylglutamic acid | + | 304.08963 | 4.66 * | 2.85 * |
| 4 | 5.7 | 1.96 | C11H15N5O3S | 5’-Methylthioadenosine | + | 297.08932 | 1.974 * | 3.32 * |
| 5 | 16.9 | 1.94 | C21H39O9P | Dolichyl b-d-glucosyl phosphate | + | 466.2366 | 2.264 * | 1.028 |
| 6 | 8.37 | 2.04 | C17H20N4O6 | Riboflavin | + | 376.13806 | 5.77 * | 1.91 * |
| 7 | 10.51 | 3.59 | C11H20O6 | Prenyl glucoside | + | 248.12555 | 2.69 * | 1.04 |
| 8 | 5.21 | 2.29 | C10H9NO | Indole acetaldehyde | + | 159.06776 | 0.17 * | 0.69 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.-Z.; Lu, X.-R.; Yang, Y.-J.; Liu, X.-W.; Qin, Z.; Li, J.-Y. Cellular Metabolomics Reveal the Mechanism Underlying the Anti-Atherosclerotic Effects of Aspirin Eugenol Ester on Vascular Endothelial Dysfunction. Int. J. Mol. Sci. 2019, 20, 3165. https://doi.org/10.3390/ijms20133165
Huang M-Z, Lu X-R, Yang Y-J, Liu X-W, Qin Z, Li J-Y. Cellular Metabolomics Reveal the Mechanism Underlying the Anti-Atherosclerotic Effects of Aspirin Eugenol Ester on Vascular Endothelial Dysfunction. International Journal of Molecular Sciences. 2019; 20(13):3165. https://doi.org/10.3390/ijms20133165
Chicago/Turabian StyleHuang, Mei-Zhou, Xiao-Rong Lu, Ya-Jun Yang, Xi-Wang Liu, Zhe Qin, and Jian-Yong Li. 2019. "Cellular Metabolomics Reveal the Mechanism Underlying the Anti-Atherosclerotic Effects of Aspirin Eugenol Ester on Vascular Endothelial Dysfunction" International Journal of Molecular Sciences 20, no. 13: 3165. https://doi.org/10.3390/ijms20133165





