In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Mice
3.2. Plasmid Construction
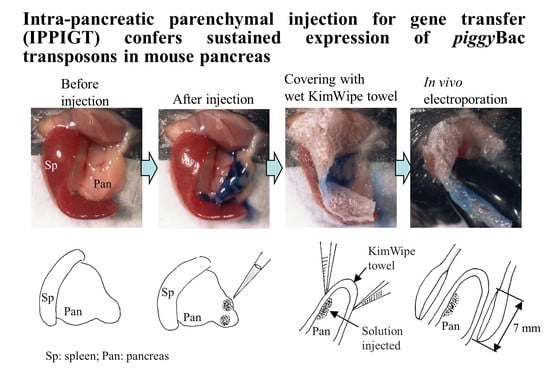
3.3. IPPIGT
3.4. Observation of Fluorescence and Dissection of Tissues
3.5. Data Analysis
3.6. Tissue Processing
3.7. PCR
3.8. Genomic Integration Site Analysis Using Splinkerette-PCR
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated viruses |
| α-GalT | α-1,3-galactosyltransferase |
| CAG | Chicken β-actin gene-based promoter |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats/CRISPR associated proteins |
| DAPI | 4′,6-diamidino-2-phenylindole |
| EGFP | Enhanced green fluorescent protein |
| EP | Electroporation |
| ES | Embryonic stem |
| FVIII | Factor VIII |
| GOI | Gene of interest |
| HGD | Hydrodynamics-based gene delivery |
| IP | Intraperitoneal |
| iPS | Induced pluripotent stem |
| IPPIGT | Intra-pancreatic parenchymal injection for gene transfer |
| ITR | Inverted terminal repeats |
| PB | PiggyBac |
| PBS | Dulbecco’s modified Ca2+, Mg2+-free phosphate-buffered saline |
| Pp | Poring pulse |
| PVP | Ployvinylpyrrolidone |
| RNAi | RNA interference |
| TB | Trypan blue |
| tdTomato | Tandem dimer Tomato |
| Tp | Transfer pulse |
References
- Manohar, M.; Verma, A.K.; Venkateshaiah, S.U.; Sanders, N.L.; Mishra, A. Pathogenic mechanisms of pancreatitis. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.M.; Weidenbach, H.; Yamagushi, H.; Lührs, H.; Liptay, S.; Adler, G. Direct gene transfer into the rat pancreas using DNA-liposomes. Eur. J. Clin. Investig. 1998, 28, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.Y.; Lin, B.; Zhang, Z.L.; Guo, L.H. Direct transfer of A20 gene into pancreas protected mice from streptozotocin- induced diabetes. Acta Pharmacol. Sin. 2004, 25, 721–726. [Google Scholar] [PubMed]
- Raper, S.E.; DeMatteo, R.P. Adenovirus-mediated in vivo gene transfer and expression in normal rat pancreas. Pancreas 1996, 12, 401–410. [Google Scholar] [CrossRef] [PubMed]
- McClane, S.J.; Chirmule, N.; Burke, C.V.; Raper, S.E. Characterization of the immune response after local delivery of recombinant adenovirus in murine pancreas and successful strategies for readministration. Hum. Gene Ther. 1997, 8, 2207–2216. [Google Scholar] [CrossRef]
- McClane, S.J.; Hamilton, T.E.; DeMatteo, R.P.; Burke, C.; Raper, S.E. Effect of adenoviral early genes and the host immune system on in vivo pancreatic gene transfer in the mouse. Pancreas 1997, 15, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, A.L.; Auricchio, A.; Yu, Q.C.; Wilson, J.; Raper, S.E. Adenoviral vector-mediated insulin gene transfer in the mouse pancreas corrects streptozotocin-induced hyperglycemia. Gene Ther. 2001, 8, 1480–1489. [Google Scholar] [CrossRef]
- Ayuso, E.; Chillón, M.; Agudo, J.; Haurigot, V.; Bosch, A.; Carretero, A.; Otaegui, P.J.; Bosch, F. In vivo gene transfer to pancreatic beta cells by systemic delivery of adenoviral vectors. Hum. Gene Ther. 2004, 15, 805–812. [Google Scholar] [CrossRef]
- Wang, A.Y.; Peng, P.D.; Ehrhardt, A.; Storm, T.A.; Kay, M.A. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Hum. Gene Ther. 2004, 15, 405–413. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, T.; Rehman, K.K.; Bertera, S.; Zhang, J.; Chen, C.; Papworth, G.; Watkins, S.; Trucco, M.; Robbins, P.D.; et al. Widespread and stable pancreatic gene transfer by adeno-associated virus vectors via different routes. Diabetes 2006, 55, 875–884. [Google Scholar] [CrossRef]
- Ayuso, E.; Chillón, M.; García, F.; Agudo, J.; Andaluz, A.; Carretero, A.; Monfar, M.; Moya, M.; Montané, J.; Otaegui, P.J.; et al. In vivo gene transfer to healthy and diabetic canine pancreas. Mol. Ther. 2006, 13, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Lafitte, M.; Rousseau, B.; Moranvillier, I.; Taillepierre, M.; Peuchant, E.; Guyonnet-Dupérat, V.; Bedel, A.; Dubus, P.; de Verneuil, H.; Moreau-Gaudry, F.; et al. In vivo gene transfer targeting in pancreatic adenocarcinoma with cell surface antigens. Mol. Cancer 2012, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, Y.; Matsuzaki, T.; Yokoo-Sugawara, M.; Suzuki, T.; Aoki, T.; Hagiwara, H.; Takata, H. Introduction and expression of glucose transporters in pancreatic acinar cells by in vivo electroporation. Acta Histochem. Cytochem. 2003, 36, 77–82. [Google Scholar] [CrossRef]
- Houbracken, I.; Baeyens, L.; Ravassard, P.; Heimberg, H.; Bouwens, L. Gene delivery to pancreatic exocrine cells in vivo and in vitro. BMC Biotechnol. 2012, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Inada, E.; Saitoh, I.; Ohtsuka, M.; Nakamura, S.; Sakurai, T.; Watanabe, S. Site-targeted non-viral gene delivery by direct DNA injection into the pancreatic parenchyma and subsequent in vivo electroporation in mice. Biotechnol. J. 2013, 8, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Somiari, S.; Glasspool-Malone, J.; Drabick, J.J.; Gilbert, R.A.; Heller, R.; Jaroszeski, M.J.; Malone, R.W. Theory and in vivo application of electroporative gene delivery. Mol. Ther. 2000, 2, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Meir, Y.J.; Coates, C.J.; Handler, A.M.; Pelczar, P.; Moisyadi, S.; Kaminski, J.M. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. USA 2006, 103, 15008–15013. [Google Scholar] [CrossRef] [PubMed]
- Ivics, Z.; Li, M.A.; Mátés, L.; Boeke, J.D.; Nagy, A.; Bradley, A.; Izsvák, Z. Transposon-mediated genome manipulation in vertebrates. Nat. Meth. 2009, 6, 415–422. [Google Scholar] [CrossRef]
- Ivics, Z. Endogenous transposase source in human cells mobilizes piggyBac transposons. Mol. Ther. 2016, 24, 851–854. [Google Scholar] [CrossRef]
- Wilson, M.H.; Coates, C.J.; George, A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007, 15, 139–145. [Google Scholar] [CrossRef]
- Clark, K.J.; Carlson, D.F.; Foster, L.K.; Kong, B.W.; Foster, D.N.; Fahrenkrug, S.C. Enzymatic engineering of the porcine genome with transposons and recombinases. BMC Biotechnol. 2007, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Kahlig, K.M.; Saridey, S.K.; Kaja, A.; Daniels, M.A.; George, A.L., Jr.; Wilson, M.H. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl. Acad. Sci. USA 2010, 107, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Saadeldin, I.M.; Choi, W.J.; Lee, S.J.; Lee, W.W.; Kim, B.H.; Han, H.J.; Bang, D.H.; Lee, B.C.; Jang, G. Production of transgenic bovine cloned embryos using piggyBac transposition. J. Vet. Med. Sci. 2011, 73, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.-P.; Yang, M.-M.; Chen, Y.-L. PiggyBac transposon-mediated gene transfer in Cashmere goat fetal fibroblast cells. Biosci. Biotechnol. Biochem. 2012, 76, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhuang, Y.; Han, M.; Xu, T.; Wu, X. piggyBac as a high-capacity transgenesis and gene-therapy vector in human cells and mice. Dis. Model Mech. 2013, 6, 828–833. [Google Scholar] [CrossRef]
- Inada, E.; Saitoh, I.; Watanabe, S.; Aoki, R.; Miura, H.; Ohtsuka, M.; Murakami, T.; Sawami, T.; Yamasaki, Y.; Sato, M. PiggyBac transposon-mediated gene delivery efficiently generates stable transfectants derived from cultured primary human deciduous tooth dental pulp cells (HDDPCs) and HDDPC-derived iPS cells. Int. J. Oral Sci. 2015, 7, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Maeda, K.; Koriyama, M.; Inada, E.; Saitoh, I.; Miura, H.; Ohtsuka, M.; Nakamura, S.; Sakurai, T.; Watanabe, S.; et al. The piggyBac-based gene delivery system can confer successful production of cloned porcine blastocysts with multigene constructs. Int. J. Mol. Sci. 2016, 17, 1424. [Google Scholar] [CrossRef]
- Xie, F.; Ye, L.; Chang, J.C.; Beyer, A.I.; Wang, J.; Muench, M.O.; Kan, Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014, 24, 1526–1533. [Google Scholar] [CrossRef]
- Chen, F.; Rosiene, J.; Che, A.; Becker, A.; LoTurco, J. Tracking and transforming neocortical progenitors by CRISPR/Cas9 gene targeting and piggyBac transposase lineage labeling. Development 2015, 142, 3601–3611. [Google Scholar] [CrossRef]
- Sato, M.; Miyoshi, K.; Nakamura, S.; Ohtsuka, M.; Sakurai, T.; Watanabe, S.; Kawaguchi, H.; Tanimoto, A. Efficient generation of somatic cell nuclear transfer-competent porcine cells with mutated alleles at multiple target loci by using CRISPR/Cas9 combined with targeted toxin-based selection system. Int. J. Mol. Sci. 2017, 18, 2610. [Google Scholar] [CrossRef]
- Wang, W.; Lin, C.; Lu, D.; Ning, Z.; Cox, T.; Melvin, D.; Wang, X.; Bradley, A.; Liu, P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 9290–9295. [Google Scholar] [CrossRef] [PubMed]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Yusa, K.; Rad, R.; Takeda, J.; Bradley, A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Meth. 2009, 6, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Sung, H.K.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C.; et al. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. Rep. 2011, 7, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, X.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lin, C.; Wang, X. PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res. 2009, 37, e141. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, Z.; Zou, X.; Zeng, F.; Shi, J.; Liu, D.; Urschitz, J.; Moisyadi, S.; Li, Z. Pig transgenesis by piggyBac transposition in combination with somatic cell nuclear transfer. Transgenic Res. 2013, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zeng, F.; Meng, F.; Xu, Z.; Zhang, X.; Huang, X.; Tang, F.; Gao, W.; Shi, J.; He, X.; et al. Generation of transgenic pigs by cytoplasmic injection of piggyBac transposase-based pmGENIE-3 plasmids. Biol. Reprod. 2014, 90, 93. [Google Scholar] [CrossRef]
- Yum, S.Y.; Lee, S.J.; Kim, H.M.; Choi, W.J.; Park, J.H.; Lee, W.W.; Kim, H.S.; Kim, H.J.; Bae, S.H.; Lee, J.H.; et al. Efficient generation of transgenic cattle using the DNA transposon and their analysis by next-generation sequencing. Sci. Rep. 2016, 6, 27185. [Google Scholar] [CrossRef]
- Li, T.; Shuai, L.; Mao, J.; Wang, X.; Wang, M.; Zhang, X.; Wang, L.; Li, Y.; Li, W.; Zhou, Q. Efficient production of fluorescent transgenic rats using the piggyBac transposon. Sci. Rep. 2016, 6, 33225. [Google Scholar] [CrossRef] [PubMed]
- Saridey, S.K.; Liu, L.; Doherty, J.E.; Kaja, A.; Galvan, D.L.; Fletcher, B.S.; Wilson, M.H. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol. Ther. 2009, 17, 2115–2120. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. PiggyBac Transposon-mediated long-term gene expression in mice. Mol. Ther. 2010, 18, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.E.; Huye, L.E.; Yusa, K.; Zhou, L.; Craig, N.L.; Wilson, M.H. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 2012, 23, 311–320. [Google Scholar] [CrossRef]
- Burnight, E.R.; Staber, J.M.; Korsakov, P.; Li, X.; Brett, B.T.; Scheetz, T.E.; Craig, N.L.; McCray, P.B., Jr. A hyperactive transposase promotes persistent gene transfer of a piggyBac DNA transposon. Mol. Ther. Nucleic Acids 2012, 1, e50. [Google Scholar] [CrossRef]
- Ley, D.; Van Zwieten, R.; Puttini, S.; Iyer, P.; Cochard, A.; Mermod, N. A PiggyBac-mediated approach for muscle gene transfer or cell therapy. Stem Cell Res. 2014, 13, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Maher, B.J.; LoTurco, J.J. piggyBac transposon-mediated cellular transgenesis in mammalian forebrain by in utero electroporation. Cold Spring Harb. Protoc. 2014, 7, 741–749. [Google Scholar] [CrossRef]
- Cooney, A.L.; Singh, B.K.; Sinn, P.L. Hybrid nonviral/viral vector systems for improved piggyBac DNA transposon in vivo delivery. Mol. Ther. 2015, 23, 667–674. [Google Scholar] [CrossRef]
- Troyanovsky, B.; Bitko, V.; Pastukh, V.; Fouty, B.; Solodushko, V. The functionality of minimal piggyBac transposons in mammalian cells. Mol. Ther. Nucleic Acids 2016, 5, e369. [Google Scholar] [CrossRef]
- Smith, R.P.; Riordan, J.D.; Feddersen, C.R.; Dupuy, A.J. A hybrid adenoviral vector system achieves efficient long-term gene expression in the liver via piggyBac transposition. Hum. Gene Ther. 2015, 26, 377–385. [Google Scholar] [CrossRef]
- Nakamura, S.; Ishihara, M.; Watanabe, S.; Ando, N.; Ohtsuka, M.; Sato, M. Intravenous delivery of piggyBac transposons as a useful tool for liver-specific gene-switching. Int. J. Mol. Sci. 2018, 19, 3452. [Google Scholar] [CrossRef] [PubMed]
- Rad, R.; Rad, L.; Wang, W.; Cadinanos, J.; Vassiliou, G.; Rice, S.; Campos, L.S.; Yusa, K.; Banerjee, R.; Li, M.A.; et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science 2010, 330, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.K.; Rad, R.; Futreal, P.A.; Bradley, A.; Liu, P. Genetic screens using the piggyBac transposon. Methods 2011, 53, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Qi, X.; Du, X.; Zou, H.; Gao, F.; Feng, T.; Lu, H.; Li, S.; An, X.; Zhang, L.; et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Miura, H.; Inoko, H.; Inoue, I.; Okada, Y.; Tanaka, M.; Sato, M.; Ohtsuka, M. PiggyBac-mediated generation of stable transfectants with surface HLA expression from a small number of cells. Anal. Biochem. 2013, 437, 29–31. [Google Scholar] [CrossRef]
- Fraser, M.J.; Ciszczon, T.; Elick, T.; Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect. Mol. Biol. 1996, 5, 141–151. [Google Scholar] [CrossRef]
- Bauser, C.A.; Eick, T.A.; Fraser, M.J. Proteins from nuclear extracts of two lepidopteran cell lines recognize the ends of TTAA-specific transposons piggyBac and tagalong. Insect. Mol. Biol. 1999, 8, 223–230. [Google Scholar] [CrossRef]
- Kettlun, C.; Galvan, D.L.; George, A.L., Jr.; Kaja, A.; Wilson, M.H. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. 2011, 19, 1636–1644. [Google Scholar] [CrossRef]
- Keith, J.H.; Schaeper, C.A.; Fraser, T.S.; Fraser, M.J., Jr. Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol. Biol. 2008, 9, 73. [Google Scholar] [CrossRef]
- Potter, C.J.; Luo, L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE 2010, 5, e10168. [Google Scholar] [CrossRef]
- Liang, A.; Wang, Y.; Woodard, L.E.; Wilson, M.H.; Sharma, R.; Awasthi, Y.C.; Du, J.; Mitch, W.E.; Cheng, J. Loss of glutathione S-transferase A4 accelerates obstruction-induced tubule damage and renal fibrosis. J. Pathol. 2012, 228, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Woodard, L.E.; Liang, A.; Luo, J.; Wilson, M.H.; Mitch, W.E.; Cheng, J. Protective role of insulin-like growth factor-1 receptor in endothelial cells against unilateral ureteral obstruction-induced renal fibrosis. Am. J. Pathol. 2015, 185, 1234–1250. [Google Scholar] [CrossRef] [PubMed]
- Yant, S.R.; Meuse, L.; Chiu, W.; Ivics, Z.; Izsvak, Z.; Kay, M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000, 25, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Fujimoto, N.; Sasakawa, N.; Ohinata, Y.; Shima, M.; Yamanaka, S.; Sugimoto, M.; Hotta, A. Delivery of fulllength factor VIII using a piggyBac transposon vector to correct a mouse model of hemophilia A. PLoS ONE 2014, 9, e104957. [Google Scholar] [CrossRef] [PubMed]
- Staber, J.M.; Pollpeter, M.J.; Arensdorf, A.; Sinn, P.L.; Rutkowski, D.T.; McCray, P.B., Jr. piggyBac-mediated phenotypic correction of factor VIII deficiency. Mol. Ther. Methods Clin. Dev. 2014, 1, 14042. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, M.; Samara-Kuko, E.; Ward, N.J.; Waddington, S.N.; McVey, J.H.; Chuah, M.K.; VandenDriessche, T. Hyperactive piggyBac transposons for sustained and robust liver-targeted gene therapy. Mol. Ther. 2014, 22, 1614–1624. [Google Scholar] [CrossRef]
- Wang, H.; Mayhew, D.; Chen, X.; Johnston, M.; Mitra, R.D. “Calling cards” for DNA-binding proteins in mammalian cells. Genetics 2012, 190, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Li, M.A.; Pettitt, S.J.; Eckert, S.; Ning, Z.; Rice, S.; Cadiñanos, J.; Yusa, K.; Conte, N.; Bradley, A. The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol. Cell. Biol. 2013, 33, 1317–1330. [Google Scholar] [CrossRef]
- Niwa, H.; Yamamura, K.; Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar] [CrossRef]
- Sato, M.; Inada, E.; Saitoh, I.; Matsumoto, Y. Microbial and enzyme technology: An efficient and convenient method for MiniPrep analysis of recombinant plasmids. J. Biomed. Sci. Eng. 2014, 7, 105–107. [Google Scholar] [CrossRef][Green Version]
- Sato, M.; Ohtsuka, M.; Nakamura, S. Intraoviductal instillation of a solution as an effective route for manipulating preimplantation mammalian embryos in vivo. In New Insights into Theriogenology; InTechOpen: Rijeka, Croatia, 2018; pp. 135–150. [Google Scholar] [CrossRef]
- Splinkerette Protocol. Available online: http://www.cmhd.ca/protocols/genetrap_pdf/Splinkerette%20Protocol%20Single%20Clone.pdf (accessed on 12 September 2018).



| Samples 1 | Sequence (5′–3′) 2 | Reference 3 |
|---|---|---|
| 12-d-1 | AGCAGTTTAAAGCAGGGACTGCTCTTGGCAGTGGACCCACCCTGCACAGTACACAGGCTCCTGTCCGGTA | AC148000| Mus musculus BAC clone RP24-105F6 from chromosome y, complete sequence; Identities = 64/64 (100%) |
| 12-d-2 | ATTGACAAGCACGCTCAATGTCGAGCCCCAATCCCTCCAACGTTTCTCTTGATCCCA | AC153511| Mus musculus 10 BAC RP23-102N12 (Roswell Park Cancer Institute (C57BL/6JFemale) Mouse BAC Library) complete sequence; Identities = 50/51 (98%) |
| 12-d-3 | GCATTGACAAGCACGCATACACATACATGCACACATGCACCT | AL513354| Mouse DNA sequence from clone RP23-150J22 on chromosome 15; Identities = 35/36 (97%) |
| 12-d-4 | GGTTTCAATTTCTTTAGTATATTCAAGCTCCGTTACCAGAGACAACTTTGGAATACAGCATCTCA | AC162174| Mus musculus chromosome 1, clone RP23-306P24, complete sequence; Identities = 59/59 (100%) |
| 12-d-5 | ACGCCTTTAATCCCAGCACTCGGGAGGCAGAGACAGGCAGATTTCTGAGTTCGAGGTCAGCC | CT030190| Mouse DNA sequence from clone RP23-115D20 on chromosome 16; Identities = 56/56 (100%) |
| 12-d-6 | TAGTGGGATCCTGGCTGTCTAGACATGTACATGATGGGCACCAAGTAAACAAGATTGATAATGAGGAAGCAGAGCTGAATAATGAAAGACACCTCGCAAAATGGGGCAT | AC132104| Mus musculus BAC clone RP24-364N7 from chromosome 9, complete sequence; Identities = 103/103 (100%) |
| 12-d-7 | GAAACTTTAAGCCCTGAAAAATTGCCTGTCAGCTTGTACCCATAAAGTAGTCTGTATAT | AC153650| Mus musculus BAC clone RP24-93I18 from chromosome 1, complete sequence; Identities = 189/189 (100%) |
| 1.5-M-1 | TAGTGGGATCGCTTGGATTCCCAGAATCCCAGTTATCTCTCCCTGCTGTCTGGGTATCCCGT | AC125181| Mus musculus BAC clone RP23-324L6 from chromosome 8, complete sequence; Identities = 56/56 (100%) |
| 1.5-M-2 | GAGACCGGGGTAAGTATGACAGTCCCCAGTGTGTGCCCTTGACTGGAATGCAGGGGTGGTGAGATGGAGCGGGATGAGGAGGAGATGGCTGAGCCTCATGTGTGGAAAGCAAGGATGCAAACAGTACCCCAC | AC142244|Mus musculus chromosome 1, clone RP23-79H24, complete sequence; Identities = 126/126 (100%) |
| 1.5-M-3 | CTAGTGGGATCCCCACCAACACATAATAGCCAGGGGCAGCAGTATATCTATATCTCCTGCAGTGGTGTATGTGGGGGG | AC157809|Mus musculus chromosome 1, clone RP24-375B12, complete sequence; Identities = 70/71 (98%) |
| 1.5-M-4 | GAGACCTTGAATCTTGTTCAAAGTACCATCAAGACTGAGGCTGCTCTTCTACAACATGCACTTTGAGAAGTTCTGCATTGGAGATGCTCAAACATCTCAGTCACTAGTAGGAAAATGAAATGGTCC | AC154681|Mus musculus BAC clone RP24-314G22 from chromosome 14, complete sequence; Identities = 23/23 (100%) |
| 1.5-M-5 | AGGGTTAAGAACCTTTAGCTAGCATGGCGGCCGAAAAGAACCCGCTCCCCGCCTCCCAGGAGCTTCTGATTGGACAACCTG | AC166750| Mus musculus chromosome 8, clone RP23-9K18, complete sequence; Identities = 71/76 (93%) |
| 1.5-M-6 | CTAGGGTTAAGTTTACTCGGAATATTTCCAGGTCTCTCCT | AC161514| Mus musculus chromosome 7, clone RP23-378L12, complete sequence; Identities = 20/20 (100%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, M.; Inada, E.; Saitoh, I.; Nakamura, S.; Watanabe, S. In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest. Int. J. Mol. Sci. 2019, 20, 3116. https://doi.org/10.3390/ijms20133116
Sato M, Inada E, Saitoh I, Nakamura S, Watanabe S. In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest. International Journal of Molecular Sciences. 2019; 20(13):3116. https://doi.org/10.3390/ijms20133116
Chicago/Turabian StyleSato, Masahiro, Emi Inada, Issei Saitoh, Shingo Nakamura, and Satoshi Watanabe. 2019. "In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest" International Journal of Molecular Sciences 20, no. 13: 3116. https://doi.org/10.3390/ijms20133116
APA StyleSato, M., Inada, E., Saitoh, I., Nakamura, S., & Watanabe, S. (2019). In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest. International Journal of Molecular Sciences, 20(13), 3116. https://doi.org/10.3390/ijms20133116