1. Introduction
Bladder cancer (BC) is a common cancer worldwide, with an estimated 429,800 new cases diagnosed and 165,100 deaths occurring annually [
1]. Bladder cancer patients can be treated by radiation, surgery, chemotherapy, and other methods of treatment to improve the survival rate of patients. However, BC has a poor prognosis and a high recurrence rate after surgery, with the 5-year overall survival rate being only 50–60% [
2,
3]. Therefore, discovering new molecular biomarkers and the molecular mechanism underlying the development and progression of BC are desperately needed.
Long noncoding RNAs (lncRNAs) are a class of RNA transcripts greater than 200 nt in length without the function of protein encoding [
4]. Although only a small number of functional lncRNAs have been well characterized to date, they have been shown to regulate gene expression at various levels, including chromatin modification, transcription, and post-transcriptional processing [
5]. Increasing evidence suggests that lncRNAs play essential roles in tumor development and could potentially serve as prognostic markers and therapeutic targets [
6,
7,
8]. We found that a lncRNA-RP11-79H23.3 (RP11-79H23.3, RP11-79H), with a length of 2994 nucleotides from chromosome 8q21.13, was significantly downregulated in bladder cancer using microarray analysis. Interestingly, phosphatase and tensin homolog (PTEN) was also downregulated concomitantly with lncRNA-RP11-79H23.3 expression and the expression of RP11-79H23.3 was negatively correlated with the clinical stage of BC. However, the biological significance and exact molecular mechanism are still unknown.
MicroRNAs (miRNAs) are a class of highly conserved small noncoding RNAs with a length of 18–25 nucleotides that could inhibit protein translation or degrade target mRNAs to regulate gene expression at a post-transcriptional level [
9]. It is well known that miRNAs can act as either an oncogene or tumor suppressor in the development and progression of cancer [
10]. Recently, a new regulatory mechanism has been proposed in which RNA transcripts can crosstalk with each other by competing for shared miRNA response elements (MREs). In this case, lncRNAs may function as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thereby modulating the depression of miRNA targets and imposing an additional level of post-transcriptional regulation [
11]. Theoretically, any RNA transcript with MREs might act as a ceRNA. If the balance of the ceRNA intricate network is disturbed, it could lead to cancer. Studies have demonstrated that a dysregulated ceRNA–ceRNA interaction network could contribute to the initialization and development of a variety of cancers, including hepatocellular carcinoma, gastric cancer, colorectal cancer, and bladder cancer [
12,
13,
14,
15]. However, the molecular mechanisms and the interaction between RP11-79H23.3 and miR-107 in BC are still unclear.
PTEN is a ubiquitous tumor suppressor that plays a critical role in cell growth and proliferation, and the functional inactivation of PTEN is associated with the pathogenesis and development of many solid tumors [
16]. PTEN is a negative regulator of the PI3K/Akt signaling pathway. Calderaro et al. found that PI3K/AKT pathway activation occurred in the entire spectrum of BC regardless of stage in 129 bladder cancer samples [
17]. In addition, loss of PTEN function contributes to hyperactivation of the PI3K/AKT pathway, which could accelerate cell growth, invasion, and metastasis in BC [
18]. A previous report demonstrated that 72% of 131 muscle-invasive bladder cancer specimens revealed dysregulation of the PI3K/Akt signaling pathway [
19]. According to target prediction using microRNA.org, the 3′-untranslated region (UTR) of PTEN is complementary to the seed region of miR-107. However, whether miR-107 can target PTEN to regulate the PI3K/AKT signaling pathway in BC has not been reported so far.
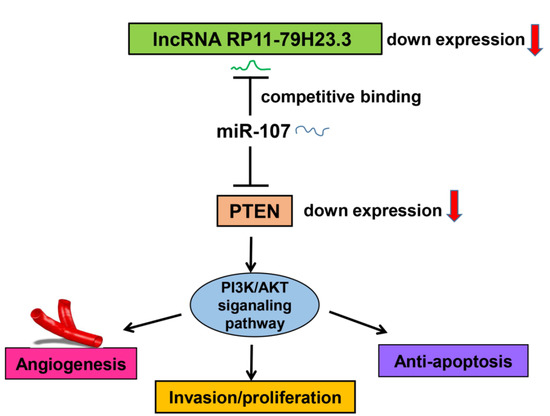
Here, we have identified a lncRNA termed lncRNA-RP11-79H23.3, the expression of which is significantly downregulated in BC tissues and cells, and the expression of RP11-79H23.3 is negatively correlated with clinical stage of BC. The present study was designed to determine whether and how RNA-RP11-79H23.3 regulates the expression of PTEN in the carcinogenesis and progression of BC. Function assays demonstrated that overexpression of RP11-79H23.3 could inhibit BC cell growth, invasion, and induce apoptosis, implying a possible role as tumor suppressor lncRNA in BC. Mechanistically, we found that RP11-79H23.3 could function as a ceRNA to regulate the target gene PTEN by sponging miR-107, which is involved in the progression of BC. Our results are of biological and clinical significance for the diagnosis and therapy of BC.
3. Discussion
It has been documented that lncRNAs play important roles in cancer development and progression, but most of them have not yet been studied in functional and mechanistic detail. In the present study, we identified a lncRNA RP11-79H23.3 which was downregulated in BC based on microarray gene-expression profile analysis. RT–qPCR analysis showed that RP11-79H23.3 levels were significantly downregulated in BC tissues and cells. We first demonstrate that RP11-79H23.3 could function as a ceRNA to regulate PTEN expression by sponging miR-107. Our data suggest that RP11-79H23.3 might play a role as a tumor suppressor in the tumorigenesis and progression of BC.
The tumor suppressor PTEN dephosphorylates the D3 position of phosphatidylinositol-3,4,5 triphosphate (PIP3) to negatively control PI3K activity and thus inhibits a panel of cellular responses mediated by the PI3K/Akt pathway including cell growth, mobility, and invasion [
20]. Tsuruta et al. found that PTEN expression was significantly reduced in bladder cancer patients, and this decrease in PTEN correlated with disease stage and grade. Thus, PTEN deficiency may contribute to initiation and progression of bladder cancer [
21]. Recently, some miRNAs have been demonstrated to promote tumorigenesis and metastasis by downregulating PTEN expression. MiR-130b could target PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway [
22]. MiR-106b and miR-93 were reported to regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer [
23]. MiR-21 promoted proliferation and invasion of triple-negative breast cancer cells through targeting PTEN [
24]. However, whether miR-107 can target PTEN in BC has not yet been reported. On the basis of bioinformatics prediction and luciferase reporter assay, we further showed miR-107 mimics significantly reduced the PTEN level, whereas miR-107 inhibitor observably enhanced PTEN expression by qRT-PCR and Western blot assays. We provided the evidence that PTEN was a direct target of miR-107 in BC.
In recent years, a ceRNA hypothesis has been proposed that RNA transcripts of coding RNAs and noncoding RNAs can communicate with each other to regulate gene expression by competing for binding to shared miRNAs [
11]. It has been reported that the abnormally expressed lncRNAs could act as ceRNAs for miRNAs to regulate the expression of miRNA target genes and disrupt the equilibrium of ceRNAs and miRNAs in tumor development. Long noncoding RNA MEG3 could function as a competing endogenous RNA for miR-181a to regulate carcinogenesis and progression of gastric cancer [
25]. LncRNA HOTAIR (HOX transcript antisense RNA) regulated HIF-1α (hypoxia inducible factor 1 subunit alpha)/AXL (AXL receptor tyrosine kinase) signaling and promoted tumorigenesis through acting as a ceRNA of miR-217 in renal cell carcinoma [
26]. Long noncoding RNA HNF1A-AS1 (HNF1A antisense RNA1) acted as a ceRNA for miR-30b-5p to promote proliferation and suppress apoptosis of bladder cancer cells through upregulating Bcl-2 [
27]. Here, we report a lncRNA, lnc-RP11-79H23.3, that might function as a tumor suppressor in human BC. The data showed that the expression of RP11-79H23.3 was positively correlated with the level of PTEN. The luciferase reporter assays found that RP11-79H23.3 and PTEN could compete for binding to miR-107. In addition, RIP assay verified that RP11-79H23.3 could directly bind to miR-107 in an AGO2-dependent manner. More importantly, RP11-79H23.3 might regulate PTEN expression by antagonizing miR-107. Together, these data confirm that RP11-79H23.3 might function as a competing endogenous RNA for miR-107 in the development of BC.
In conclusion, we found that RP11-79H23.3 was downregulated in bladder cancer and could liberate miR-107 via its function as a ceRNA to suppress PTEN expression and activate the PI3K/Akt signaling pathway, which consequently contributes to the pathogenesis and progression of BC. Taken together, our findings suggest that the RP11-79H23.3 could play a potential tumor suppressor role in the progression and development of BC. The RP11-79H23.3/miR-107/PTEN axis might serve as a novel clinical marker and therapeutic target for BC.
4. Materials and Methods
4.1. Patient and Tissue Samples
Bladder cancer tissues and pair-matched adjacent tissues used in this paper were obtained from the First Affiliated Hospital of Chongqing Medical University and the Affiliated Hospital of Southwest Medical University during 2010–2015 in accordance with the Helsinki Declaration. None of the patients had undergone radiation treatment or chemotherapy before surgery. Informed consent was obtained from these patients. Tumors were classified according to the tumor–node–metastasis (TNM, 2010) system of classification. The clinicopathological characteristics of the BC patients are summarized in
Table 1. This study was approved by the Ethics Committee of Chongqing Medical University.
4.2. Cell Lines, Plasmid Construction, and Transfection
Human normal bladder epithelial cell line (SV-HUC-1) and bladder cancer cell lines (EJ, T24, BIU87) were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were maintained in a humidified incubator at 37 °C in an atmosphere of 5% CO2. EJ, T24, BIU87 and SV-HUC-1 cells were maintained in RPMI-1640 and DMEM/F12K medium, respectively.
Full-length RP11-79H23.3 from 293 cells was amplified by PCR and then cloned into pIRES2-EGFP vector (Shanghai, China). The sequences of RP11-79H23.3 3′-UTR and the sequence of PTEN 3′-UTR containing putative miR-107 binding sites and the corresponding mutant (Mut) 3′-UTR sequences were amplified (F-RP11-79H23.3-Wt: GCGGCTCGAGGTCGTGGACTAATGAAACA, R-RP11-79H23.3-Wt: AATGCGGCCGCTACCAATGACATGAAAGGG, F-RP11-79H23.39-Mut1: TCATTCTGACGACTTAACTTCTACATGGTGC, R-RP11-79H23.39-Mut1: GAAGTTAAGTCGTCAGAATGAAACCAGACTG, F-RP11-79H23.39-Mut2: TTAATGTGACGACGATTATAACCTAGGTTATT, R-RP11-79H23.39-Mut2: GTTATAATCGTCGTCACATTAATTTCCAAATC, F-PTEN-3′UTR-Wt: GCGGCTCGAGACCAACTGAAGTGGCTAA, R-PTEN-3′UTR-Mut: ACGATCCTGTCGTCCTATTATGATTGAAAACT, F-PTEN-3′UTR-Mut: TCATAATAGGACGACAGGATGCTTCATGTGCTG, R-PTEN-3′UTR-Wt: AATGCGGCCGCAAACTCTATAAATGCTGC) and then cloned into the pmiR-RB-Report™ vector (RIBOBIO, Guangzhou, China). si-RP11-79H23.3 (sense I: 5′-GCTTCAACTCTGTGAAATA-3′, sense II: 5′-CCTATTTCTTACCATCCTT-3′) was synthesized by RIBOBIO, miR-107 mimics, miR-107 inhibitor, and negative control, which were from GenePharma (Shanghai, China). The transfections were executed with a Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s guide.
4.3. LncRNA and mRNA Microarray
Briefly, as previously reported (Huang et al., 2016 [
15]), four pairs of human BC and the adjacent normal tissues were applied for microarray assay to determine differentially expressed lncRNAs and mRNAs by the Arraystar Human lncRNA Array v2.0 (KangChen Bio-tech, Shanghai, China). The transcript of the significant differential expression was reserved by fold change ≥2.0,
p < 0.05, and FDR < 0.05. Hierarchical clustering was implemented to produce an outline of expression patterns according to the expression value of all the transcripts with or without significant differences. Microarray data were available through Gene Expression Omnibus (GEO) with accession code GSE89006.
4.4. qRT-PCR Assay
Total RNA was extracted from the frozen tissues and cultured cells with TRIzol (TAKARA, Dalian, China). The quantity and quality of RNA were detected by Nano Drop 2000 spectrophotometer (Nano Drop Thermo, Wilmington, DE, USA), and the integrity of RNA was evaluated by agarose gel electrophoresis. cDNA was generated with a reverse-transcription kit (Takara, Dalian, China) according to manufacturer’s protocol. The qRT-PCR analysis was implemented by the ABI 7900HT Sequence Detection Machine (Bio-Rad, Hercules, CA, USA) using SYBR Green chemistry. U6 or GAPDH was used as an endogenous control. The specific primers sequences were as follows: RP11-79H23.3, 5′-TGGCCTCAGTTAGGACTGCT-3′ and 5′-CTGCTTCCGCTCTCTTTCTC-3′; PTEN, 5′-GCTATGGGATTTCCTGCAGAA-3′ and 5′-GGCGGTGTCATAATGTCTTTCA-3′; GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. PCR was executed with an initial denaturation step at 95 °C for 5 min, followed by amplification with 40 cycles at 95 °C for 10 s and 60 °C for 35 s, the melt curve step at 60 °C to 95 °C, and the increment at 0.5 °C for 5 s. The genes were amplified in separate wells in triplicate. The relative expressions levels were calculated with the (2−∆∆Ct) method.
4.5. Cell Proliferation, Viability, and Colony Assays
The cell proliferation ability was determined by CCK8, EdU and colony formation assays. For CCK-8 assay, Cell Counting Kit-8 was bought from DingGuo (Beijing, China), and cells were seeded into 96-well plates (5000 cells/well) with complete growth medium. After incubation for 24, 48, 72, and 96 h, respectively, 10 μL CCK-8 was added into each well, and then the cells were cultured for an additional 2 h. The absorbance value was tested by a plate reader at 450 nm (Bio-Rad, Hercules, CA, USA). 1 × 105 cells were put into 24-well plates for EdU incorporation with an EdU detection kit (Ribobio, Guangzhou, China). The percentage of EdU-positive cells was quantified from four random fields per well. For colony formation assays, 2.5 × 102 cells, were plated onto six-well plates and incubated for 14 days, fixed with 4% paraform, then stained with 0.1% crystal violet, and the number of colonies in four random fields were counted under an inverted microscope. The experiments were performed in triplicate.
4.6. Wound Healing, Cell Invasion Assay, and Cytoskeleton
When cells grew in six-well plates to 80–90% confluence, scratches were made with a 200 μL tip, they were washed with PBS (Phosphate Buffered Saline), and then the cells were further incubated in FBS (fetal bovine serum)-free medium. The width of wound closure (original width at 0 h–width after cell migration at 24 h) was counted in the same wound point with five replicates. The cell invasion assays were performed using 24-well Transwell chambers with a pore size of 8 μm (BD BioCoat, Bedford, MA, USA). 5 × 105 cells in serum-free RPMI1640 were added to the upper chamber coated with matrigel (BD Biosciences, Franklin Lakes, NJ, USA), and the bottom chamber contained 500 µL of culture medium with 10% FBS. After 24 h, noninvading cells on the upper chamber were rubbed away with a cotton bud and invading cells in the lower chamber were stained by crystal violet, photographed, and counted with an Olympus multifunction microscope (Tokyo, Japan). Assays were repeated three times. The cells were seeded into 24-well plates after 48 h, rinsed with PBS, fixed in 4% paraformaldehyde, permeabilized by 0.3% Triton X-100 at 37 °C for 6 min, then rinsed three times with PBS, and blocked with 3% BSA (bovine serum albumin) at 37 °C for 30 min. The cells were incubated with Paxillin antibody (1:200 dilution) overnight at 4 °C, then incubated with IgG-TRITC for 1 h, then incubated with 1% FITC-phalloidin (Sigma Chemical Corp., St. Louis, MO, USA) at 37 °C for 30 min. The cells were stained by DAPI (4′,6-diamidino-2-phenylindole, SouthernBiotech, Birmingham, AL, USA) for 10 min, sealed with antifluorescent solution, and were then were observed under a confocal laser scanning microscope (Leica, Wetzlar, Germany).
4.7. Cell Apoptosis and Cell Cycle Assay
The cells were fixed with 4% paraform for 20 min, then dyed with Hoechst 33342 for 15 min and observed under a Leica TCS-SP2 Laser Confocal Microscopy. Apoptosis was assessed by TUNEL assay using an apoptosis detection kit (Keygen Biotech, Nanjing, China). The FITC-labeled apoptotic cells were photographed under a fluorescent microscope. The cells were harvested and apoptosis was determined with flow cytometry (Becon Dickinson FACSCalibur, New York, NY, USA) after annexin V-FITC/PI double staining. 2 × 106 cells were fixed for 12 h in 70% ethanol, then stained with PI. The cell cycle was observed with flow cytometry (Becon Dickinson FACSCalibur, New York, NY, USA).
4.8. Fluorescence In Situ Hybridization (FISH)
The subcellular localizations of LncRNA RP11-79H23.3 and miR-107 were examined by FISH kit (Roche Applied Science, Penzberg, Germany). Cells grew to 70% confluence, were fixed with 4% formaldehyde for 15 min, then permeabilized with 0.5% TritonX-100 for 15 min and rinsed with PBS. Cells were incubated in a mixture of RP11-79H23.3 probes labeled with Cy3 at 37 °C overnight and washed with prewarmed 2× saline-sodium citrate (SSC). The FITC-labeled miR-107 probes were incubated in prehybridization buffer (1:100) at 88 °C for 5 min and at 4 °C for 3 min with the PCR instrument (Bio-Rad, Hercules, CA, USA). Next, the FITC-labeled miR-107 probes were added into cells at 37 °C overnight, washed with 2× SSC, and stained by DAPI. Observations were undertaken with a Leica TCS-SP2 Laser Scanning Confocal Microscope.
4.9. Dual-Luciferase Reporter Assay
Pmir-RB-RP11-79H23.3(RP11-79H-wt), pmir-RB-RP11-79H23.3-mutant (RP11-79H-mut), pmir-RB-PTEN (PTEN 3′-UTR-wt), or pmir-RB-PTEN-mutant (PTEN 3′-UTR-mut) was co-transfected using Lipofectamine 2000 with miR-107 mimic or NC-mimic and miR-107 inhibitor or NC-inhibitor or pIRES2-RP11-79H23.3 into EJ cells. According to the manufacturer’s protocol, luciferase activities were determined using the dual-luciferase assay kit (Promega, Madison, WI, USA) after 48 h of transfection.
4.10. RIP (RNA Immunoprecipitation) Assay
RIP is an antibody-based technique used to map in vivo RNA-protein interactions. The RNA binding protein (RBP) of interest is immunoprecipitated together with its associated RNA for identification of bound transcripts (mRNAs, noncoding RNAs, or viral RNAs). Transcripts are detected by real-time PCR. RIP detection was implemented according to the manufacturer’s guide with the Magna RNA binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). EJ cells were transfected with miR-107 mimics and miR-107 NC, respectively. Then, after cells were lysed by RIP lysis buffer, whole cell lysate was incubated in the buffer containing anti-AGO2 antibody coupled to magnetic beads (Millipore). RNA was separated and deposited by immunoprecipitation, then the products were purified and analyzed by qRT-PCR. In addition, EJ cells were co-transfected with pcDNA3.1-MS2, pcDNA3.1-MS2-RB-RP11-79H23.3, and pcDNA3.1-MS2-RB-RP11-79H23.3-mut (no miR-107 binding site), respectively, and pMS2-GFP (Addgene plasmids). After 48 h, RIP experiments were executed using an anti-GFP antibody (Abcam, Burlingame, CA, USA) with IgG antibody as a negative antibody. The products were purified and enriched to detect the target miRNAs by qRT-PCR.
4.11. Western Blot Analysis
Cell protein lysates were extracted from cells using 1× sodium dodecyl sulfate buffer and quantified by Bicin Choninic Acid (BCA) protein assay kit (Thermo, Waltham, MA, USA). On a 10% SDS-PAGE (Sodium doecylsulfate-polyacrylamide gel electrophoresis), 30 μg of protein samples per lane were separated and then electransferred to a PVDF (polyvinylidene fluoride) membrane. The samples were blocked in 5% nonfat milk powder and then incubated with antibodies against PTEN (Abcam, Burlingame, CA, USA), AKT, p-AKT, PI3K, p-PI3K,GSK3β, p-GSK3β, Bax, Bcl-2, Caspase-3 cleaved (Cell Signaling Technology, Beverly, MA, USA), and β-actin (Proteintech, Rosemont, IL, USA) at 4 °C overnight. Blots were incubated with goat anti-rabbit secondary antibodies (Bioss, Beijing, China) for 2 h at room temperature. β-actin was used as an internal control. The bands were visualized by an ECL chemiluminescent detection system (Thermo Scientific, New York, NY, USA).
4.12. Immunohistochemistry (IHC) and Immunofluorescence (IF)
Cells were cultured on the slides in a 24-well plate and fixed. The frozen and paraffin section were obtained from nude mouse tumor tissues and human tissues for IF and IHC, respectively. For IHC assay, after dewaxing, rehydration, and antigen retrieval, the slides were then incubated with antibodies specific for PTEN and PI3K/AKT signaling pathway molecules (Abcam) and secondary HRP labelled goat anti-rabbit, stained with DAB, then counterstained with hematoxylin. For IF assays, the cells and frozen section were incubated with indicated primary antibodies overnight at 4 °C. Then, samples were incubated with fluorescein Alexa-Fluor 488- or 594-conjugated secondary antibodies. The slides were dyed by DAPI, then observed under a confocal laser microscope and Olympus multifunction microscope.
4.13. Tumor Xenograft Model
BALB/c nude mice (4–6 weeks old, female) were purchased from Beijing HFK Bioscience Co., Ltd. (Beijing, China). The stably transfected EJ cells (2 × 106) with indicated vector or 3′-cholesterol and 2′-OMe-conjugated siRNA and NC random fragments (RIBOBIO) were subcutaneously injected into BALB/c mice, respectively. The volume of tumor was measured per week. The mice were sacrificed after 35 days, and tumor xenografts and lungs were harvested and subjected to pathologic, immunohistochemical, and immunofluorescent examination. The microvessels were counted from six different fields in the tumor HE sections under microscope. Metastatic nodules of the lung were recorded under the microscope. All animal care and experiment procedures were implemented according to the guidelines of the National Institutes of Health. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Chongqing Medical University (Date: 16 February 2016, Identification code: 2016021606).
4.14. Statistical Analysis
Statistical analyses were executed by Student’s t-test and One-way ANOVA using GraphPad Prism 6.0 and SPSS 19.0 statistical software. The significance of differences between groups was assessed by Student’s t test and Fisher’s exact test. The data were expressed as mean ± SD. p < 0.05 was considered as statistical significance.