Systemic Activation of Activin A Signaling Causes Chronic Kidney Disease-Mineral Bone Disorder
Abstract
:1. Introduction
2. Regulation of Osteoclastic Development and Bone Resorption
3. Activin a Biology in Osteoclastogenesis
4. Regulation of Osteogenesis and Bone Formation
5. Activin a Biology in Osteoblast Development and Function
6. The Mechanisms of the Medial Vascular Calcification Caused by CKD
7. Activin a Biology in CKD-Induced Medial Vascular Calcification
8. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Vale, W.; Rivier, J.; Vaughan, J.; McClintock, R.; Corrigan, A.; Woo, W.; Karr, D.; Spiess, J. Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature 1986, 321, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Ying, S.-Y.; Ueno, N.; Shimasaki, S.; Esch, F.; Hotta, M.; Guillemin, R. Pituitary FSH is released by a heterodimer of the β subunits from the two forms of inhibin. Obstet. Gynecol. Surv. 1987, 42, 109–111. [Google Scholar] [CrossRef]
- Mathews, L.S. Activin receptors and cellular signaling by the receptor serine kinase family. Endocr. Rev. 1994, 15, 310–325. [Google Scholar] [CrossRef] [PubMed]
- Hedger, M.P.; de Kretser, D.M. The activins and their binding protein, follistatin—Diagnostic and therapeutic targets in inflammatory disease and fibrosis. Cytokine Growth Factor Rev. 2013, 24, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy—From molecular mechanisms to therapeutic benefits. Biochin. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Zarubin, T.; Han, J. Activation and signaling of thep38 MAP kinase pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Mansell, A.; Patella, S.; Scott, B.J.; Hedger, M.P.; de Kretser, D.M.; Phillips, D.J. Activin A is a critical component of the inflammatory response, and its binding protein, follistatin, reduces mortality in endotoxemia. Proc. Natl. Acad. Sci. USA 2007, 104, 16239–16244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauklin, S.; Vallier, L. Activin/Nodal signalling in stem cells. Development 2015, 142, 607–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijayarathna, R.; de Kretser, D.M. Activins in reproductive biology and beyond. Hum. Reprod. Update 2016, 22, 342–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, K.; Funaba, M. Activin in humoral immune responses. Vitam. Horm. 2011, 85, 235–253. [Google Scholar] [PubMed]
- Hedger, M.P.; Winnall, W.R.; Phillips, D.J.; de Kretser, D.M. The regulation and functions of activin and follistatin in inflammation and immunity. Vitam. Horm. 2011, 85, 255–297. [Google Scholar] [PubMed]
- Antsiferova, M.; Werner, S. The bright and the dark sides of activin in wound healing and cancer. J. Cell Sci. 2012, 125, 3929–3937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antsiferova, M.; Martin, C.; Huber, M.; Feyerabend, T.B.; Förster, A.; Hartmann, K.; Rodewald, H.R.; Hohl, D.; Werner, S. Mast cells are dispensable for normal and activin-promoted wound healing and skin carcinogenesis. J. Immunol. 2013, 191, 6147–6155. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.D.; Yu, R.T.; Kakizuka, A.; Kintner, C.R.; Mathews, L.S.; Vale, W.W.; Evans, R.M.; Umesono, K. Activin and its receptors during gastrulation and the later phases of mesoderm development in the chick embryo. Dev. Biol. 1995, 172, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Ball, E.M.; Risbridger, G.P. Activins as regulators of branching morphogenesis. Dev. Biol. 2001, 238, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Merino, R.; Macias, D.; Gañan, Y.; Rodriguez-Leon, J.; Economides, A.N.; Rodriguez-Esteban, C.; Izpisua-Belmonte, J.C.; Hurle, J.M. Control of digit formation by activin signalling. Development 1999, 126, 2161–2170. [Google Scholar] [PubMed]
- Matzuk, M.M.; Kumar, T.R.; Vassalli, A.; Bickenbach, J.R.; Roop, D.R.; Jaenisch, R.; Bradley, A. Functional analysis of activins during mammalian development. Nature 1995, 374, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Matzuk, M.M.; Kumar, T.R.; Shou, W.; Coerver, K.A.; Lau, A.L.; Behringer, R.R.; Finegold, M.J. Transgenic models to study the roles of inhibins and activins in reproduction, oncogenesis, and development. Recent Prog. Horm. Res. 1996, 51, 123–154. [Google Scholar] [PubMed]
- Brown, C.W.; Houston-Hawkins, D.E.; Woodruff, T.K.; Matzuk, M.M. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat. Genet. 2000, 25, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Perry, K.W.; Salusky, I.B. Chronic kidney disease mineral and bone disorder. In Endocrinology: Adult and Pediatric, 7th ed.; Elsevier: Philadelphia, PA, USA, 2016; pp. 1214–1229. [Google Scholar]
- Cunningham, J.; Sprague, S.; Cannata-Andia, J.; Coco, M.; Cohen-Solal, M.; Fitzpatrick, L.; Goltzmann, D.; Lafage-Proust, M.H.; Leonard, M.; Ott, S.; et al. Osteoporosis in chronic kidney disease. Am. J. Kidney Dis. 2004, 43, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Agapova, O.A.; Fang, Y.; Sugatani, T.; Seifert, M.E.; Hruska, K.A. Ligand trap for the activin type IIA receptor protects against vascular disease and renal fibrosis in mice with chronic kidney disease. Kidney Int. 2016, 89, 1231–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugatani, T.; Agapova, O.A.; Fang, Y.; Berman, A.G.; Wallace, J.M.; Malluche, H.H.; Faugere, M.C.; Smith, W.; Sung, V.; Hruska, K.A. Ligand trap of the activin receptor type IIA inhibits osteoclast stimulation of bone remodeling in diabetic mice with chronic kidney disease. Kidney Int. 2017, 91, 86–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.J.; Sugatani, T.; Agapova, O.A.; Fang, Y.; Gaut, J.P.; Faugere, M.C.; Malluche, H.H.; Hruska, K.A. The activin receptor is stimulated in the skeleton, vasculature, heart, and kidney during chronic kidney disease. Kidney Int. 2018, 93, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Nakashima, T.; Shinohara, M.; Negishi-Koga, T.; Komatsu, N.; Terashima, A.; Sawa, S.; Nitta, T.; Takayanagi, H. Osteoimmunology: The Conceptual Framework Unifying the Immune and Skeletal Systems. Physiol. Rev. 2017, 97, 1295–1349. [Google Scholar] [CrossRef] [PubMed]
- Janckila, A.J.; Takahashi, K.; Sun, S.Z.; Yam, L.T. Tartrate-resistant acid phosphatase isoform 5b as serum marker for osteoclastic activity. Clin. Chem. 2001, 47, 74–80. [Google Scholar] [PubMed]
- Dougall, W.C.; Glaccum, M.; Charrier, K.; Rohrbach, K.; Brasel, K.; De Smedt, T.; Daro, E.; Smith, J.; Tometsko, M.E.; Maliszewski, C.R.; et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999, 13, 2412–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Y.Y.; Yoshida, H.; Sarosi, I.; Tan, H.L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.; Lacey, D.L.; Dunstan, C.R.; Solovyev, I.; Colombero, A.; Timms, E.; Tan, H.L.; Elliott, G.; Kelley, M.J.; Sarosi, I.; et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 1999, 96, 3540–3545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, N.; Kinosaki, M.; Yamaguchi, K.; Shima, N.; Yasuda, H.; Yano, K.; Morinaga, T.; Higashio, K. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem. Biophys. Res. Commun. 1998, 253, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamana, H.; Yoshiki, S.; Roodman, G.D.; Mundy, G.R.; Jones, S.J.; Boyde, A.; Suda, T. Osteoclast-like cell formation and its regulation by osteotropic hormones in mouse bone marrow cultures. Endocrinology 1988, 122, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Suda, T.; Takahashi, N.; Martin, T.J. Modulation of osteoclast differentiation. Endocr. Rev. 1992, 13, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogianni, G.; Mann, V.; Noble, B.S. Apoptotic bodies convey activity capable of initiating osteoclastogenesis and localized bone destruction. J. Bone Miner. Res. 2008, 23, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef]
- Bucay, N.; Sarosi, I.; Dunstan, C.R.; Morony, S.; Tarpley, J.; Capparelli, C.; Scully, S.; Tan, H.L.; Xu, W.; Lacey, D.L.; et al. Osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998, 12, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, A.; Amizuka, N.; Irie, K.; Murakami, A.; Fujise, N.; Kanno, T.; Sato, Y.; Nakagawa, N.; Yasuda, H.; Mochizuki, S.; et al. Severe osteoporosis in mice lacking osteoclastogenesis inhibitory factor/osteoprotegerin. Biochem. Biophys. Res. Commun. 1998, 247, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Lüthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef]
- Tondravi, M.M.; McKercher, S.R.; Anderson, K.; Erdmann, J.M.; Quiroz, M.; Maki, R.; Teitelbaum, S.L. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature 1997, 386, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Weilbaecher, K.N.; Motyckova, G.; Huber, W.E.; Takemoto, C.M.; Hemesath, T.J.; Xu, Y.; Hershey, C.L.; Dowland, N.R.; Wells, A.G.; Fisher, D.E. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol. Cell 2001, 8, 49–58. [Google Scholar] [CrossRef]
- Lagasse, E.; Weissman, I.L. Enforced expression of Bcl-2 in monocytes rescues macrophages and partially reverses osteopetrosis in op/op mice. Cell 1997, 89, 1021–1031. [Google Scholar] [CrossRef]
- Takayanagi, H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Inui, M.; Inoue, K.; Kim, S.; Suematsu, A.; Kobayashi, E.; Iwata, T.; Ohnishi, H.; Matozaki, T.; Kodama, T.; et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 2004, 428, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Takami, M.; Rho, J.; Josien, R.; Choi, Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J. Exp. Med. 2002, 195, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Cella, M.; Buonsanti, C.; Strader, C.; Kondo, T.; Salmaggi, A.; Colonna, M. Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J. Exp. Med. 2003, 198, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H. Osteoimmunology and the effects of the immune system on bone. Nat. Rev. Rheumatol. 2009, 5, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Balkan, W.; Martinez, A.F.; Fernandez, I.; Rodriguez, M.A.; Pang, M.; Troen, B.R. Identification of NFAT binding sites that mediate stimulation of cathepsin K promoter activity by RANK ligand. Gene 2009, 446, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Crotti, T.N.; Sharma, S.M.; Fleming, J.D.; Flannery, M.R.; Ostrowski, M.C.; Goldring, S.R.; McHugh, K.P. PU.1 and NFATc1 mediate osteoclastic induction of the mouse beta3 integrin promoter. J. Cell. Physiol. 2008, 215, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, S.H.; Ha Kim, J.; Choi, Y.; Kim, N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol. Endocrinol. 2008, 22, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Yoneda, T.; Lowe, C.; Soriano, P.; Mundy, G.R. Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J. Clin. Investig. 1992, 90, 1622–1627. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, I.; Gailit, J.; Sasaki, T. Osteoclast integrin alphaVbeta3 is present in the clear zone and contributes to cellular polarization. Cell Tissue Res. 1996, 286, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Biskobing, D.M.; Fan, D. Acid pH increases carbonic anhydrase II and calcitonin receptor mRNA expression in mature osteoclasts. Calcif. Tissue Int. 2000, 67, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Teitelbaum, S.L.; Ghiselli, R.; Gluck, S. Osteoclastic bone resorption by a polarized vacuolar proton pump. Science 1989, 245, 855–857. [Google Scholar] [CrossRef] [PubMed]
- Kornak, U.; Kasper, D.; Bösl, M.R.; Kaiser, E.; Schweizer, M.; Schulz, A.; Friedrich, W.; Delling, G.; Jentsch, T.J. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 2001, 104, 205–215. [Google Scholar] [CrossRef]
- Wilson, S.R.; Peters, C.; Saftig, P.; Brömme, D. Cathepsin K activity-dependent regulation of osteoclast actin ring formation and bone resorption. J. Biol. Chem. 2009, 284, 2584–2592. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Nomura, S.; Hosoi, T.; Ouchi, Y.; Orimo, H.; Muramatsu, M. Localization of follistatin, an activin-binding protein, in bone tissues. Calcif. Tissue Int. 1994, 55, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Hosoi, T.; Inoue, S.; Hoshino, S.; Ouchi, Y.; Orimo, H. Immunohistochemical detection of activin A in osteoclasts. Gerontology 1996, 42, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Lotinun, S.; Pearsall, R.S.; Horne, W.C.; Baron, R. Activin receptor signaling: A potential therapeutic target for osteoporosis. Curr. Mol. Pharmacol. 2012, 5, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shao, L.E.; Lemas, V.; Yu, A.L.; Vaughan, J.; Rivier, J.; Vale, W. Importance of FSH-releasing protein and inhibin in erythrodifferentiation. Nature 1987, 330, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Kawakita, M.; Kato, K.; Yonemura, Y.; Masuda, T.; Matsuzaki, H.; Hirose, J.; Isaji, M.; Sasaki, H.; Inoue, T.; et al. Purification of megakaryocyte differentiation activity from a human fibrous histiocytoma cell line: N-terminal sequence homology with activin, A. Biochem. Biophys. Res. Commun. 1991, 174, 1163–1168. [Google Scholar] [CrossRef]
- Okafuji, K.; Kaku, K.; Seguchi, M.; Tanaka, H.; Azuno, Y.; Kaneko, T. Effects of activin A/erythroid differentiation factor on erythroid and megakaryocytic differentiations of mouse erythroleukemia (Friend) cells: Evidence for two distinct modes of cell response. Exp. Hematol. 1995, 23, 210–216. [Google Scholar] [PubMed]
- Broxmeyer, H.E.; Lu, L.; Cooper, S.; Schwall, R.H.; Mason, A.J.; Nikolics, K. Selective and indirect modulation of human multipotential and erythroid hematopoietic progenitor cell proliferation by recombinant human activin and inhibin. Proc. Natl. Acad. Sci. USA 1988, 85, 9052–9056. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Suzuki, T.; Hashimoto, M.; Eto, Y.; Shiokawa, K.; Muramatsu, M. Induction of differentiation of the human promyelocytic cell line HL-60 by activin/EDF. Biochem. Biophys. Res. Commun. 1992, 187, 79–85. [Google Scholar] [CrossRef]
- Fuller, K.; Bayley, K.E.; Chambers, T.J. Activin A is an essential cofactor for osteoclast induction. Biochem. Biophys. Res. Commun. 2000, 268, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Gaddy-Kurten, D.; Coker, J.K.; Abe, E.; Jilka, R.L.; Manolagas, S.C. Inhibin suppresses and activin stimulates osteoblastogenesis and osteoclastogenesis in murine bone marrow cultures. Endocrinology 2002, 143, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Perrien, D.S.; Achenbach, S.J.; Bledsoe, S.E.; Walser, B.; Suva, L.J.; Khosla, S.; Gaddy, D. Bone turnover across the menopause transition: Correlations with inhibins and follicle-stimulating hormone. J. Clin. Endocrinol. Metab. 2006, 9, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Sakai, R.; Eto, Y.; Ohtsuka, M.; Hirafuji, M.; Shinoda, H. Activin enhances osteoclast-like cell formation in vitro. Biochem. Biophys. Res. Commun. 1993, 195, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Murase, Y.; Okahashi, N.; Koseki, T.; Itoh, K.; Udagawa, N.; Hashimoto, O.; Sugino, H.; Noguchi, T.; Nishihara, T. Possible involvement of protein kinases and Smad2 signaling pathways on osteoclast differentiation enhanced by activin, A. J. Cell. Physiol. 2001, 188, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Alvarez, U.M.; Hruska, K.A. Activin A stimulates IkappaB-alpha/NFkappaB and RANK expression for osteoclast differentiation, but not AKT survival pathway in osteoclast precursors. J. Cell. Biochem. 2003, 90, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Silbermann, R.; Bolzoni, M.; Storti, P.; Guasco, D.; Bonomini, S.; Zhou, D.; Wu, J.; Anderson, J.L.; Windle, J.J.; Aversa, F.; et al. Bone marrow monocyte-/macrophage-derived activin A mediates the osteoclastogenic effect of IL-3 in multiple myeloma. Leukemia 2014, 28, 951–954. [Google Scholar] [CrossRef] [PubMed]
- Pearsall, R.S.; Canalis, E.; Cornwall-Brady, M.; Underwood, K.W.; Haigis, B.; Ucran, J.; Kumar, R.; Pobre, E.; Grinberg, A.; Werner, E.D.; et al. A soluble activin type IIA receptor induces bone formation and improves skeletal integrity. Proc. Natl. Acad. Sci. USA 2008, 105, 7082–7087. [Google Scholar] [CrossRef] [PubMed]
- Lotinun, S.; Pearsall, R.S.; Davies, M.V.; Marvell, T.H.; Monnell, T.E.; Ucran, J.; Fajardo, R.J.; Kumar, R.; Underwood, K.W.; Seehra, J.; et al. A soluble activin receptor Type IIA fusion protein (ACE-011) increases bone mass via a dual anabolic-antiresorptive effect in Cynomolgus monkeys. Bone 2010, 46, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Ruckle, J.; Jacobs, M.; Kramer, W.; Pearsall, A.E.; Kumar, R.; Underwood, K.W.; Seehra, J.; Yang, Y.; Condon, C.H.; Sherman, M.L. Single-dose, randomized, double-blind, placebo-controlled study of ACE-011 (ActRIIA-IgG1) in postmenopausal women. J. Bone Miner. Res. 2009, 24, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Borton, A.J.; Frederick, J.P.; Datto, M.B.; Wang, X.F.; Weinstein, R.S. The loss of Smad3 results in a lower rate of bone formation and osteopenia through dysregulation of osteoblast differentiation and apoptosis. J. Bone Miner. Res. 2001, 16, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Omata, Y.; Yasui, T.; Hirose, J.; Izawa, N.; Imai, Y.; Matsumoto, T.; Masuda, H.; Tokuyama, N.; Nakamura, S.; Tsutsumi, S.; et al. Genomewide comprehensive analysis reveals critical cooperation between Smad and c-Fos in RANKL-induced osteoclastogenesis. J. Bone Miner. Res. 2015, 30, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Kajita, T.; Ariyoshi, W.; Okinaga, T.; Mitsugi, S.; Tominaga, K.; Nishihara, T. Mechanisms involved in enhancement of osteoclast formation by activin-A. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Goto, D.; Hamamoto, T.; Takenoshita, S.; Kato, M.; Miyazono, K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 1999, 274, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Johnson, K.; Chen, H.J.; Carroll, S.; Laughon, A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 1997, 388, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Y.F.; Jayaraman, L.; Yang, H.; Massague, J.; Pavletich, N.P. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGF-beta signaling. Cell 1998, 94, 585–594. [Google Scholar] [CrossRef]
- Kawabata, M.; Inoue, H.; Hanyu, A.; Imamura, T.; Miyazono, K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998, 17, 4056–4065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennler, S.; Huet, S.; Gauthier, J.M. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene 1999, 18, 1643–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massague, J.; Wotton, D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000, 19, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Ten Dijke, P.; Hill, C.S. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 2004, 29, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Dunn, N.R.; Koonce, C.H.; Anderson, D.C.; Islam, A.; Bikoff, E.K.; Robertson, E.J. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes Dev. 2005, 19, 152–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, K.A.; Pietenpol, J.A.; Moses, H.L. A tale of two proteins: Differential roles and regulation of Smad2 and Smad3 in TGF-β signaling. J. Cell. Biochem. 2007, 101, 9–33. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Liang, Y.Y.; Liang, M.; Zhai, W.; Lin, X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 2002, 9, 133–143. [Google Scholar] [CrossRef]
- Alliston, T.; Ko, T.C.; Cao, Y.; Liang, Y.Y.; Feng, X.H.; Chang, C.; Derynck, R. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J. Biol. Chem. 2005, 280, 24227–24237. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Weisberg, E.; Fridmacher, V.; Watanabe, M.; Naco, G.; Whitman, M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 1997, 389, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Labbe, E.; Silvestri, C.; Hoodless, P.A.; Wrana, J.L.; Attisano, L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 1998, 2, 109–120. [Google Scholar] [CrossRef]
- Liu, F.; Massague, J.; Altaba, A.R. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat. Genet. 1998, 20, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.M.; Williams, M.E.; Heaton, J.H.; Gelehrter, T.D.; Innis, J.W. Group 13 HOX proteins interact with the MH2 domain of R-Smads and modulate Smad transcriptional activation functions independent of HOX DNA-binding capability. Nucleic Acids Res. 2005, 33, 4475–4484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellemans, J.; Preobrazhenska, O.; Willaert, A.; Debeer, P.; Verdonk, P.C.; Costa, T.; Janssens, K.; Menten, B.; Roy, N.V.; Vermeulen, S.J.; et al. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 2004, 36, 1213–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, Z.A.; Yang, C.C.; Wrana, J.L.; McDermott, J.C. Smad proteins function as co-modulators for MEF2 transcriptional regulatory proteins. Nucleic Acids Res. 2001, 29, 732–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanai, J.; Chen, L.F.; Kanno, T.; Ohtani-Fujita, N.; Kim, W.Y.; Guo, W.H.; Imamura, T.; Ishidou, Y.; Fukuchi, M.; Shi, M.J.; et al. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J. Biol. Chem. 1999, 274, 31577–31582. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Lin, X.; Derynck, R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 2000, 19, 5178–5193. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.A. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003, 22, 2443–2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.T.; Yamaguchi, S.; Hirano, K.; Ichisaka, T.; Kuroda, T.; Tada, T. Nanog co-regulated by Nodal/Smad2 and Oct4 is required for pluripotency in developing mouse epiblast. Dev. Biol. 2014, 392, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Dankbar, B.; Fennen, M.; Brunert, D.; Hayer, S.; Frank, S.; Wehmeyer, C.; Beckmann, D.; Paruzel, P.; Bertrand, J.; Redlich, K.; et al. Myostatin is a direct regulator of osteoclast differentiation and its inhibition reduces inflammatory joint destruction in mice. Nat. Med. 2015, 21, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Ovitt, C.; Grigoriadis, A.E.; Mohle-Steinlein, U.; Ruther, U.; Wagner, E.F. Bone and haematopoietic defects in mice lacking c-fos. Nature 1992, 360, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Owens, J.M.; Tonko, M.; Elliott, C.; Chambers, T.J.; Wagner, E.F. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 2000, 24, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Galson, D.L.; Zhao, C.; Peng, L.; Laplace, C.; Wang, K.Z.; Bachler, M.A.; Amano, H.; Aburatani, H.; Ishikawa, H.; et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 2004, 279, 26475–26480. [Google Scholar] [CrossRef] [PubMed]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Tu, A.W.; Luo, K. Acetylation of Smad2 by the co-activator p300 regulates activin and transforming growth factor beta response. J. Biol. Chem. 2007, 282, 21187–21196. [Google Scholar] [CrossRef] [PubMed]
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011, 13, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, H.M. Developmental regulation of the growth plate. Nature 2003, 423, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Tang, Y.; Wu, X.; Lei, W.; Pang, L.; Wan, C.; Shi, Z.; Zhao, L.; Nagy, T.R.; Peng, X.; Hu, J.; et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009, 15, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Kaji, H.; Naito, J.; Sowa, H.; Sugimoto, T.; Chihara, K. Smad3 differently affects osteoblast differentiation depending upon its differentiation stage. Horm. Metab. Res. 2006, 38, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Alliston, T.; Choy, L.; Ducy, P.; Karsenty, G.; Derynck, R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001, 20, 2254–2272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjelmeland, A.B.; Schilling, S.H.; Guo, X.; Quarles, D.; Wang, X.F. Loss of Smad3-mediated negative regulation of Runx2 activity leads to an alteration in cell fate determination. Mol. Cell. Biol. 2005, 25, 9460–9468. [Google Scholar] [CrossRef] [PubMed]
- Centrella, M.; McCarthy, T.L.; Canalis, E. Activin-A binding and biochemical effects in osteoblast-enriched cultures from fetal-rat parietal bone. Mol. Cell. Biol. 1991, 11, 25025–25028. [Google Scholar] [CrossRef]
- Hashimoto, M.; Shoda, A.; Inoue, S.; Yamada, R.; Kondo, T.; Sakurai, T.; Ueno, N.; Muramatsu, M. Functional regulation of osteoblastic cells by the interaction of activin-A with follistatin. J. Biol. Chem. 1992, 267, 4999–5004. [Google Scholar] [PubMed]
- Oue, Y.; Kanatani, H.; Kiyoki, M.; Eto, Y.; Ogata, E.; Matsumoto, T. Effect of local injection of activin A on bone formation in newborn rats. Bone 1994, 15, 361–366. [Google Scholar] [CrossRef]
- Ikenoue, T.; Jingushi, S.; Urabe, K.; Okazaki, K.; Iwamoto, Y. Inhibitory effects of activin-A on osteoblast differentiation during cultures of fetal rat calvarial cells. J. Cell. Biochem. 1999, 75, 206–214. [Google Scholar] [CrossRef]
- Rosenberg, N.; Soudry, M.; Rosenberg, O.; Blumenfeld, I.; Blumenfeld, Z. The role of activin A in the human osteoblast cell cycle: A preliminary experimental in vitro study. Exp. Clin. Endocrinol. Diabetes 2010, 118, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Chantry, A.D.; Heath, D.; Mulivor, A.W.; Pearsall, S.; Baud’huin, M.; Coulton, L.; Evans, H.; Abdul, N.; Werner, E.D.; Bouxsein, M.L.; et al. Inhibiting activin-A signaling stimulates bone formation and prevents cancer-induced bone destruction in vivo. J. Bone Miner. Res. 2010, 25, 2633–2646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loots, G.G.; Keller, H.; Leupin, O.; Murugesh, D.; Collette, N.M.; Genetos, D.C. TGF-β regulates sclerostin expression via the ECR5 enhancer. Bone 2012, 50, 663–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, R.D.; Eijken, M.; Bezstarosti, K.; Demmers, J.A.; van Leeuwen, J.P. Activin A suppresses osteoblast mineralization capacity by altering extracellular matrix (ECM) composition and impairing matrix vesicle (MV) production. Mol. Cell. Proteom. 2013, 12, 2890–2900. [Google Scholar] [CrossRef] [PubMed]
- Hruska, K.A.; Seifert, M.; Sugatani, T. Pathophysiology of the chronic kidney disease-mineral bone disorder. Curr. Opin. Nephrol. Hypertens. 2015, 24, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Hruska, K.A.; Sugatani, T.; Agapova, O.; Fang, Y. The chronic kidney disease—Mineral bone disorder (CKD-MBD): Advances in pathophysiology. Bone 2017, 100, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Byon, C.H.; Yuan, K.; Chen, J.; Mao, X.; Heath, J.M.; Javed, A.; Zhang, K.; Anderson, P.G.; Chen, Y. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ. Res. 2012, 111, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Giachelli, C.M. Vascular calcification in CKD-MBD: Roles for phosphate, FGF23, and Klotho. Bone 2017, 100, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.L.; Behrmann, A.; Shao, J.S.; Ramachandran, B.; Krchma, K.; Bello Arredondo, Y.; Kovacs, A.; Mead, M.; Maxson, R.; Towler, D.A. Targeted reduction of vascular Msx1 and Msx2 mitigates arteriosclerotic calcification and aortic stiffness in LDLR-deficient mice fed diabetogenic diets. Diabetes 2014, 63, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Masuda, M.; Miyazaki-Anzai, S.; Keenan, A.L.; Shiozaki, Y.; Okamura, K.; Chick, W.S.; Williams, K.; Zhao, X.; Rahman, S.M.; Tintut, Y.; et al. Activating transcription factor-4 promotes mineralization in vascular smooth muscle cells. JCI Insight 2016, 1, e88646. [Google Scholar] [CrossRef] [PubMed]
- Du, K.L.; Ip, H.S.; Li, J.; Chen, M.; Dandre, F.; Yu, W.; Lu, M.M.; Owens, G.K.; Parmacek, M.S. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 2003, 23, 2425–2437. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Sato, H.; Doi, H.; Yoshida, C.A.; Shimizu, T.; Matsui, H.; Yamazaki, M.; Akiyama, H.; Kawai-Kowase, K.; Iso, T.; et al. Runx2 represses myocardin-mediated differentiation and facilitates osteogenic conversion of vascular smooth muscle cells. Mol. Cell. Biol. 2008, 28, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.E.; Chen, T.; Leaf, E.M.; Speer, M.Y.; Giachelli, C.M. Runx2 Expression in Smooth Muscle Cells Is Required for Arterial Medial Calcification in Mice. Am. J. Pathol. 2015, 185, 1958–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaz, U.; Schellinger, I.N.; Chernogubova, E.; Warnecke, C.; Kayama, Y.; Penov, K.; Hennigs, J.K.; Salomons, F.; Eken, S.; Emrich, F.C.; et al. Transcription Factor Runx2 Promotes Aortic Fibrosis and Stiffness in Type 2 Diabetes Mellitus. Circ. Res. 2015, 117, 513–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Matsuda, K.; Bialek, P.; Jacquot, S.; Masuoka, H.C.; Schinke, T.; Li, L.; Brancorsini, S.; Sassone-Corsi, P.; Townes, T.M.; et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 2004, 117, 387–398. [Google Scholar] [CrossRef]
- Yang, X.; Karsenty, G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J. Biol. Chem. 2004, 279, 47109–47114. [Google Scholar] [CrossRef] [PubMed]
- Clark-Greuel, J.N.; Connolly, J.M.; Sorichillo, E.; Narula, N.R.; Rapoport, H.S.; Mohler, E.R.; Gorman, J.H.; Gorman, R.C.; Levy, R.J. Transforming growth factor-β1 mechanisms in aortic valve calcification: Increased alkaline phosphatase and related events. Ann. Thorac. Surg. 2007, 83, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Narula, N.; Li, Q.Y.; Mohler, E.R.; Levy, R.J. Progression of aortic valve stenosis: TGF-β1 is present in calcified aortic valve cusps and promotes aortic valve interstitial cell calcification via apoptosis. Ann. Thorac. Surg. 2003, 75, 457–465. [Google Scholar] [CrossRef]
- Anastasilakis, A.D.; Polyzos, S.A.; Makras, P.; Gkiomisi, A.; Savvides, M.; Papatheodorou, A.; Terpos, E. Circulating activin-A is elevated in postmenopausal women with low bone mass: The three-month effect of zoledronic acid treatment. Osteoporos. Int. 2013, 24, 2127–2132. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Shintani, Y.; Sakamoto, Y.; Wakatsuki, M.; Shitsukaw, K.; SaitoSerum, S. Immunoreactive activin A levels in normal subjects and patients with various diseases. J. Clin. Endocrinol. Metab. 1996, 81, 2125–2130. [Google Scholar] [PubMed]
- Wakatsuki, M.; Shintani, Y.; Abe, M.; Liu, Z.H.; Shitsukawa, K.; Saito, S. Immunoradiometric assay for follistatin: Serum immunoreactive follistatin levels in normal adults and pregnant women. J. Clin. Endocrinol. Metab. 1996, 81, 630–634. [Google Scholar] [PubMed]
- Loria, P.; Petraglia, F.; Concari, M.; Bertolotti, M.; Martella, P.; Luisi, S.; Grisolia, C.; Foresta, C.; Volpe, A.; Genazzani, A.R.; et al. Influence of age and sex on serum concentrations of total dimeric activin, A. Eur. J. Endocrinol. 1998, 139, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Ueland, T.; Aukrust, P.; Aakhus, S.; Smith, C.; Endresen, K.; Birkeland, K.I.; Gullestad, L.; Johansen, O.E. Activin A and cardiovascular disease in type 2 diabetes mellitus. Diabetes Vasc. Dis. Res. 2012, 9, 234–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, H.J.; Huber, T.B.; Isermann, B.; Schiffer, M. CKD in diabetes: Diabetic kidney disease versus nondiabetic kidney disease. Nat. Rev. Nephrol. 2018, 14, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Kooman, J.P.; Dekker, M.J.; Usvyat, L.A.; Kotanko, P.; van der Sande, F.M.; Schalkwijk, C.G.; Shiels, P.G.; Stenvinkel, P. Inflammation and premature aging in advanced chronic kidney disease. Am. J. Physiol. Renal Physiol. 2017, 313, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, C.M. Mechanism of vascular calcification in CKD-evidence for premature ageing? Nat. Rev. Nephrol. 2013, 9, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Fordyce, C.; Fessenden, T.; Pickering, C.; Jung, J.; Singla, V.; Berman, H.; Tlsty, T. DNA damage drives an activin a-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev. Res. 2010, 3, 190–201. [Google Scholar] [CrossRef] [PubMed]
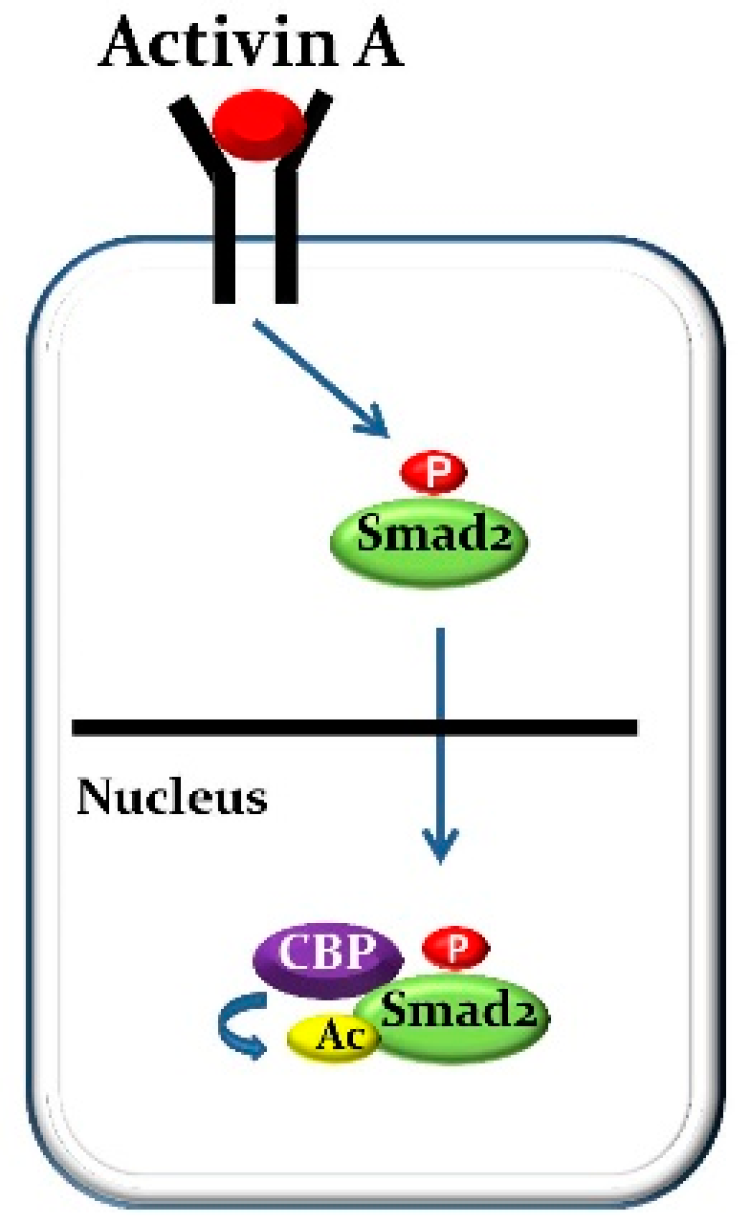
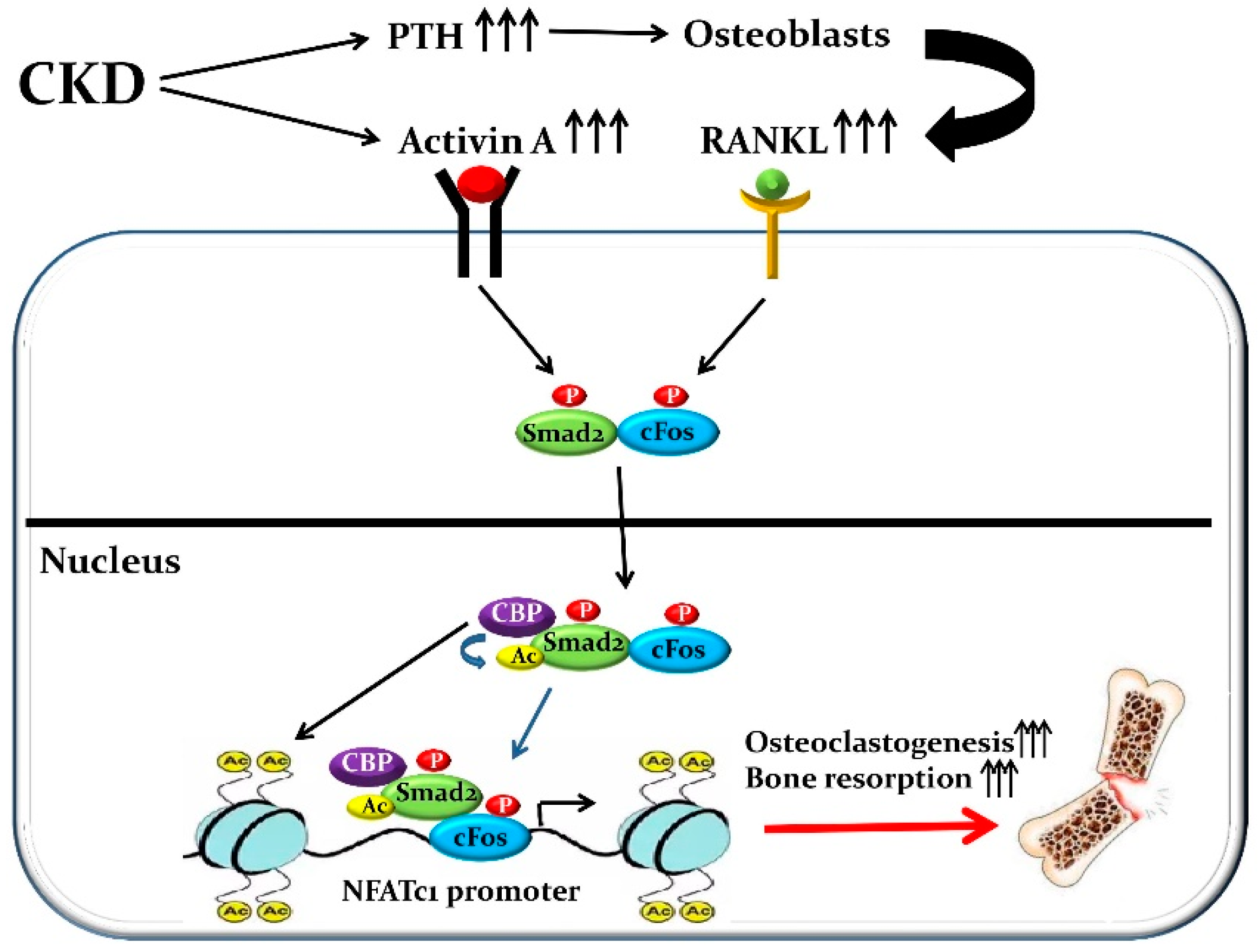
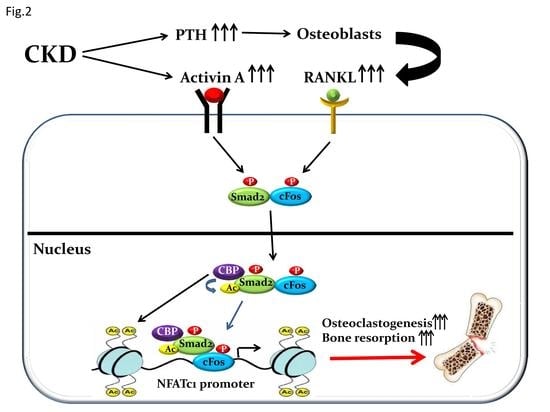
| Transcription Factors: Official Symbol/(Official Full Name) | References |
|---|---|
| Fos (FBJ osteosarcoma oncogene) | [24,76,77] |
| Myc (MYC proto-oncogene, bHLH transcription factor) | [87] |
| Evi-1 (ecotropic viral integration site 1) | [88] |
| Foxh1 (forkhead box H1) | [89,90] |
| Gli3 (GLI-Kruppel family member GLI3) | [91] |
| Hoxa13 (homeobox A13) | [92] |
| Lemd3 (LEM domain containing 3) | [93] |
| Mef2a (myocyte enhancer factor 2A) | [94] |
| Runx2 (runt related transcription factor 2) | [95] |
| Sp1 (trans-acting transcription factor 1) | [96] |
| Zeb1 (zinc finger E-box binding homeobox 1) | [97] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugatani, T. Systemic Activation of Activin A Signaling Causes Chronic Kidney Disease-Mineral Bone Disorder. Int. J. Mol. Sci. 2018, 19, 2490. https://doi.org/10.3390/ijms19092490
Sugatani T. Systemic Activation of Activin A Signaling Causes Chronic Kidney Disease-Mineral Bone Disorder. International Journal of Molecular Sciences. 2018; 19(9):2490. https://doi.org/10.3390/ijms19092490
Chicago/Turabian StyleSugatani, Toshifumi. 2018. "Systemic Activation of Activin A Signaling Causes Chronic Kidney Disease-Mineral Bone Disorder" International Journal of Molecular Sciences 19, no. 9: 2490. https://doi.org/10.3390/ijms19092490