The Protein Phosphatase PPM1G Destabilizes HIF-1α Expression
Abstract
1. Introduction
2. Results
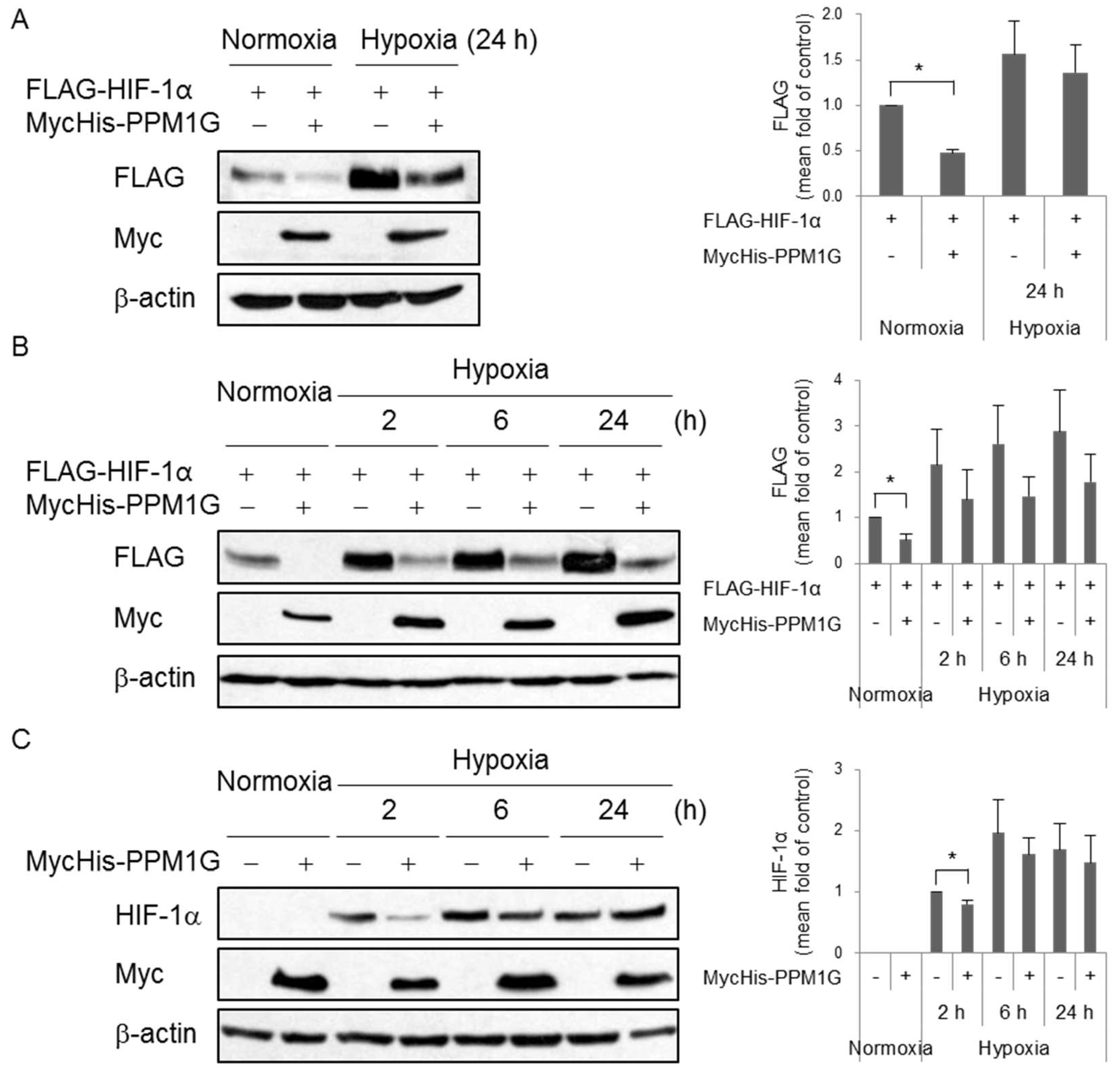
2.1. Hypoxia-Inducible Factor (HIF)-1α Expression Is Negatively Regulated by Protein Phosphatase 1 Gamma (PPM1G)
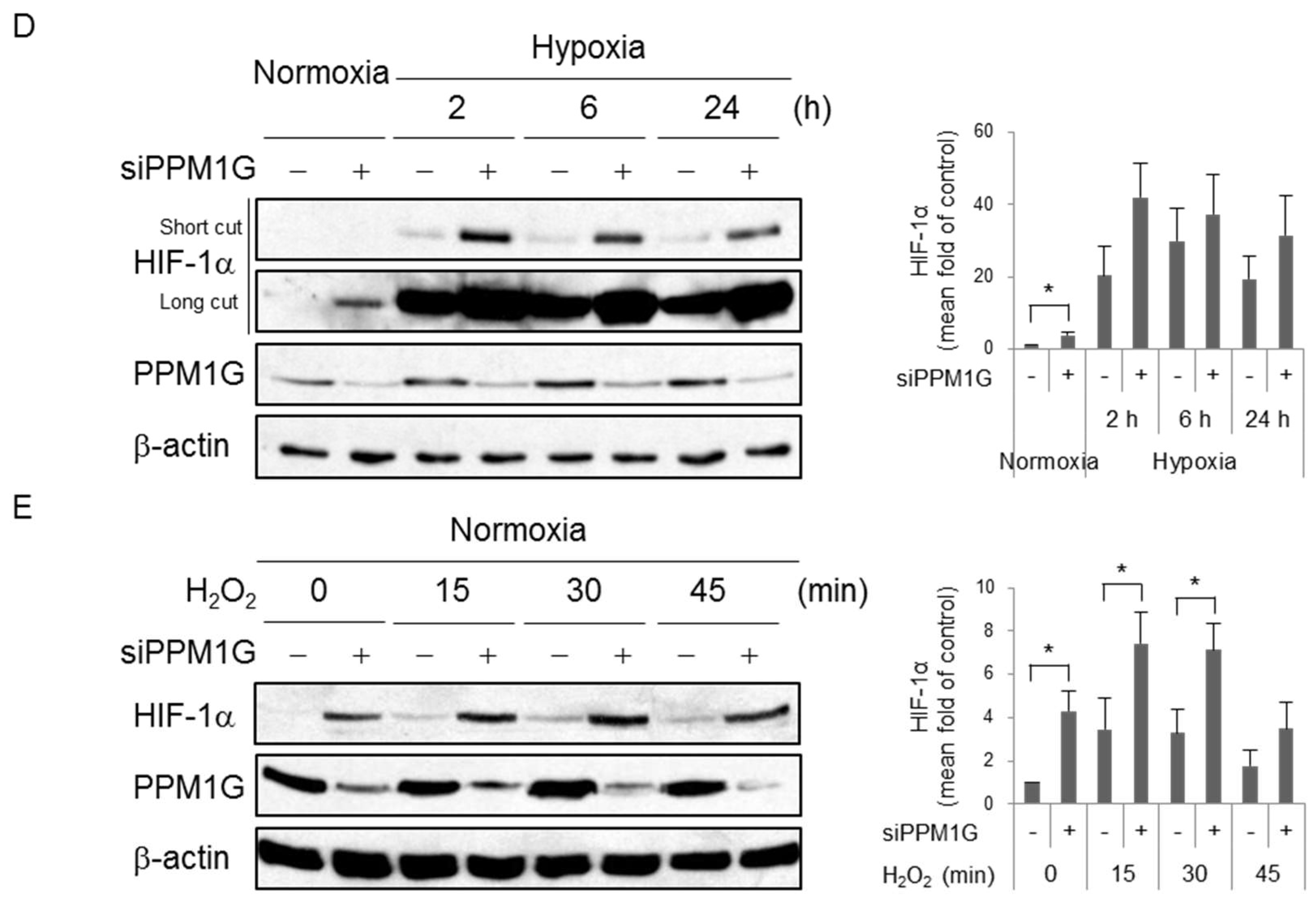
2.2. HIF-1α Is Downregulated by PPM1G in Normoxia and Upon Acute Hypoxic and Oxidative Stress
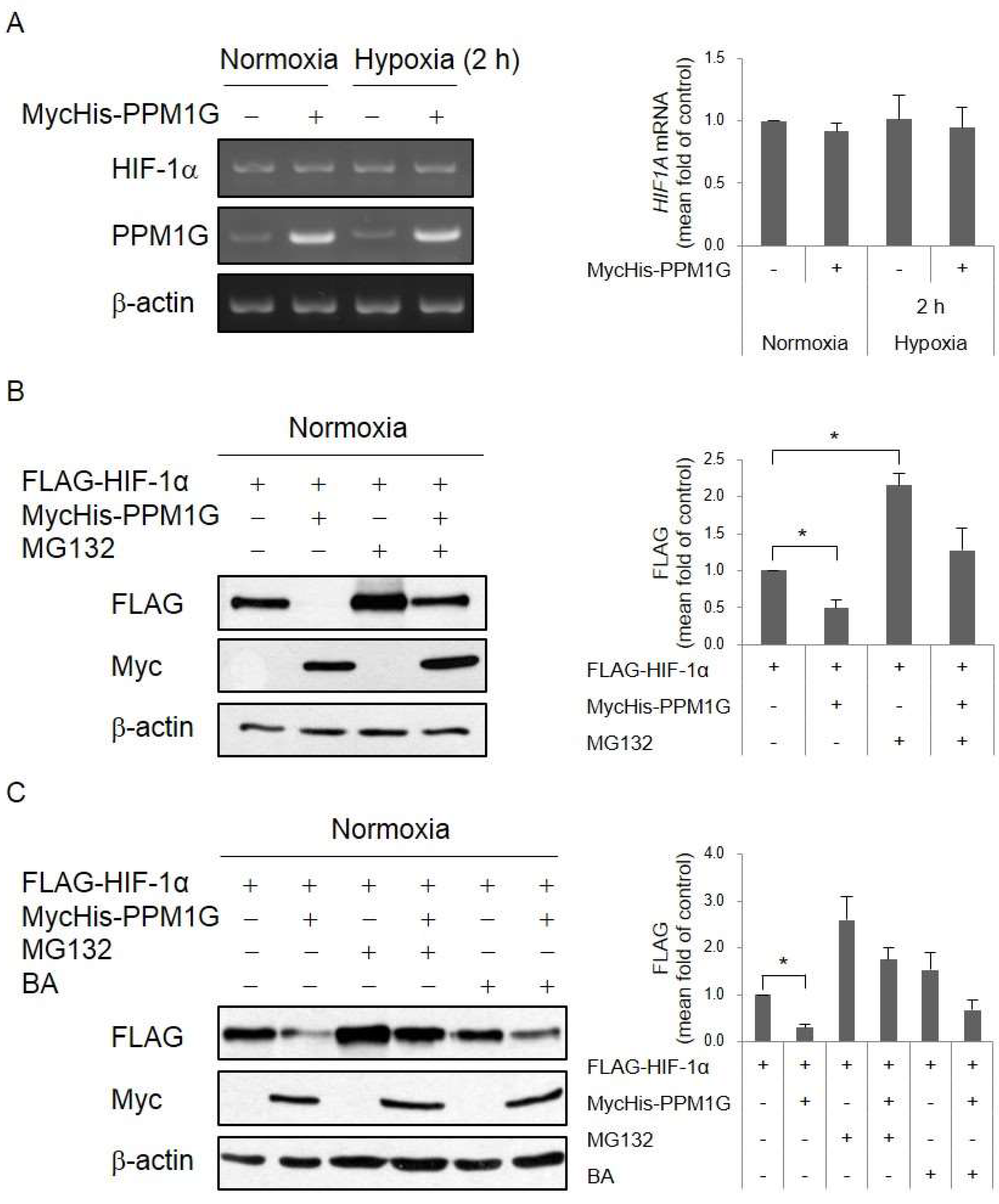
2.3. PPM1G Promotes HIF-1α Degradation via the Proteasomal Pathway
2.4. PPM1G Promotes HIF-1α Degradation in a PHD-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Chemical Compounds
4.2. Cell Culture, Hypoxia Condition and Treatment
4.3. Plasmid Transfection
4.4. Small Interfering RNA (siRNA) Transfection
4.5. Preparation of Crude Cell Extract and Western Blotting
4.6. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.7. Luciferase Reporter Assay
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ARNT | Aryl hydrocarbon receptor nuclear translocator |
| BA | Bafilomycin A1 |
| CCAR2 | Cell cycle and apoptosis regulator 2 |
| DMOG | Dimethyloxaloylglycine |
| HIF | Hypoxia-inducible factor |
| HRE | Hypoxia response elements |
| PCAF | p300/CBP-associated factor |
| PHD | Prolyl-4-hydroxylase domain-containing proteins |
| PPM1G | Mg2+/Mn2+-dependent protein phosphatase 1 gamma |
| VHL | Von Hippel-Lindau |
References
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef]
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Wenger, R.H.; Stiehl, D.P.; Camenisch, G. Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005, 2005, re12. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Whitelaw, M.L.; Peet, D.J. The hypoxia-inducible factors: Key transcriptional regulators of hypoxic responses. Cell. Mol. Life Sci. 2003, 60, 1376–1393. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shen, S.M.; Zhao, X.Y.; Chen, G.Q. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int. J. Biochem. Mol. Biol. 2012, 3, 165–178. [Google Scholar] [PubMed]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.E.; Gu, J.; Schau, M.; Bunn, H.F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7987–7992. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFαtargeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Berta, M.A.; Mazure, N.; Hattab, M.; Pouyssegur, J.; Brahimi-Horn, M.C. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 2007, 360, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.H.; Jeong, J.W.; Park, J.A.; Kim, S.H.; Bae, M.K.; Choi, S.J.; Kim, K.W. Sumoylation increases HIF-1αstability and its transcriptional activity. Biochem. Biophys. Res. Commun. 2004, 324, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Carbia-Nagashima, A.; Gerez, J.; Perez-Castro, C.; Paez-Pereda, M.; Silberstein, S.; Stalla, G.K.; Holsboer, F.; Arzt, E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1α during hypoxia. Cell 2007, 131, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kang, X.; Zhang, S.; Yeh, E.T. SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 2007, 131, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Li, J.; Zou, Y.; Yi, J.; Zhang, H.; Cao, M.; Yeh, E.T.; Cheng, J. PIASy stimulates HIF1α SUMOylation and negatively regulates HIF1α activity in response to hypoxia. Oncogene 2010, 29, 5568–5578. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Liu, Q.; Xue, C.; David, L.L.; Beer, T.M.; Thomas, G.V.; Dai, M.S.; Qian, D.Z. HIF1α protein stability is increased by acetylation at lysine 709. J. Biol. Chem. 2012, 287, 35496–35505. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Bae, M.K.; Ahn, M.Y.; Kim, S.H.; Sohn, T.K.; Bae, M.H.; Yoo, M.A.; Song, E.J.; Lee, K.J.; Kim, K.W. Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef]
- Kim, Y.; Nam, H.J.; Lee, J.; Park, D.Y.; Kim, C.; Yu, Y.S.; Kim, D.; Park, S.W.; Bhin, J.; Hwang, D.; et al. Methylation-dependent regulation of HIF-1α stability restricts retinal and tumour angiogenesis. Nat. Commun. 2016, 7, 10347. [Google Scholar] [CrossRef] [PubMed]
- Mennerich, D.; Dimova, E.Y.; Kietzmann, T. Direct phosphorylation events involved in HIF-α regulation: The role of GSK-3β. Hypoxia (Auckl.) 2014, 2, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Mennerich, D.; Dimova, E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Flugel, D.; Gorlach, A.; Kietzmann, T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 2012, 119, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yao, Y.; Lu, L.; Costa, M.; Dai, W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1α. J. Biol. Chem. 2010, 285, 38944–38950. [Google Scholar] [CrossRef] [PubMed]
- Warfel, N.A.; Dolloff, N.G.; Dicker, D.T.; Malysz, J.; El-Deiry, W.S. CDK1 stabilizes HIF-1α via direct phosphorylation of Ser668 to promote tumor growth. Cell Cycle 2013, 12, 3689–3701. [Google Scholar] [CrossRef] [PubMed]
- Cam, H.; Easton, J.B.; High, A.; Houghton, P.J. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol. Cell. 2010, 40, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Schober, A.S.; Berra, E. DUBs, New Members in the Hypoxia Signaling clUb. Front. Oncol. 2016, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Jiao, H.; Wang, W.; Mei, Z.; Chen, G. Sumoylation of hypoxia inducible factor-1α and its significance in cancer. Sci. China Life Sci. 2014, 57, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.Y.; Yun, M.; Jeong, J.; Park, E.R.; Shin, H.J.; Woo, S.R.; Jung, J.K.; Kim, Y.M.; Park, J.J.; Kim, J.; et al. SIRT1 deacetylates and stabilizes hypoxia-inducible factor-1α (HIF-1α) via direct interactions during hypoxia. Biochem. Biophys. Res. Commun. 2015, 462, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.Z.; Kachhap, S.K.; Collis, S.J.; Verheul, H.M.; Carducci, M.A.; Atadja, P.; Pili, R. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 α. Cancer Res. 2006, 66, 8814–8821. [Google Scholar] [CrossRef] [PubMed]
- Alig, S.K.; Stampnik, Y.; Pircher, J.; Rotter, R.; Gaitzsch, E.; Ribeiro, A.; Wornle, M.; Krotz, F.; Mannell, H. The tyrosine phosphatase SHP-1 regulates hypoxia inducible factor-1α (HIF-1α) protein levels in endothelial cells under hypoxia. PLoS ONE 2015, 10, e0121113. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, Y.; Du, Y.; Ning, X.; Liu, N.; Huang, D.; Liang, J.; Xue, Y.; Fan, D. Dual-specificity phosphatase DUSP1 protects overactivation of hypoxia-inducible factor 1 through inactivating ERK MAPK. Exp. Cell Res. 2005, 309, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yogesha, S.D.; Mayfield, J.E.; Gill, G.N.; Zhang, Y. Viewing serine/threonine protein phosphatases through the eyes of drug designers. FEBS J. 2013, 280, 4739–4760. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.V.; Kobayashi, R.; Krainer, A.R. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999, 13, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Allemand, E.; Hastings, M.L.; Murray, M.V.; Myers, M.P.; Krainer, A.R. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 2007, 14, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Petri, S.; Grimmler, M.; Over, S.; Fischer, U.; Gruss, O.J. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J. Cell Biol. 2007, 179, 451–465. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.P.; McCann, J.L.; Gudipaty, S.A.; D’Orso, I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013, 5, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Gudipaty, S.A.; McNamara, R.P.; Morton, E.L.; D’Orso, I. PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation. Mol. Cell. Biol. 2015, 35, 3810–3828. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stevens, P.D.; Eshleman, N.E.; Gao, T. Protein phosphatase PPM1G regulates protein translation and cell growth by dephosphorylating 4E binding protein 1 (4E-BP1). J. Biol. Chem. 2013, 288, 23225–23233. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wang, L.; Feng, W.; Feng, Y.; Shu, H.K. Phosphatidylinositol-3 kinase-dependent translational regulation of Id1 involves the PPM1G phosphatase. Oncogene 2016, 35, 5807–5816. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, G.; Wrighton, K.H.; Lin, H.; Songyang, Z.; Feng, X.H.; Lin, X. Regulation of p27Kip1 phosphorylation and G1 cell cycle progression by protein phosphatase PPM1G. Am. J. Cancer Res. 2016, 6, 2207–2220. [Google Scholar] [PubMed]
- Suh, E.J.; Kim, T.Y.; Kim, S.H. PP2Cgamma-mediated S-phase accumulation induced by the proteasome-dependent degradation of p21(WAF1/CIP1). FEBS Lett. 2006, 580, 6100–6104. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Takizawa, N.; Allemand, E.; Hori, T.; Iborra, F.J.; Nozaki, N.; Muraki, M.; Hagiwara, M.; Krainer, A.R.; Fukagawa, T.; et al. A novel histone exchange factor, protein phosphatase 2Cgamma, mediates the exchange and dephosphorylation of H2A-H2B. J. Cell Biol. 2006, 175, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 2012, 46, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Khoronenkova, S.V.; Dianova, I.I.; Ternette, N.; Kessler, B.M.; Parsons, J.L.; Dianov, G.L. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol. Cell 2012, 45, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Foster, W.H.; Langenbacher, A.; Gao, C.; Chen, J.; Wang, Y. Nuclear phosphatase PPM1G in cellular survival and neural development. Dev. Dyn. 2013, 242, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.N.; Yang, W.K.; Kim, J.; Kim, H.S.; Kim, E.J.; Yun, H.; Park, H.; Kim, S.S.; Choe, W.; Kang, I.; et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 α protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis 2008, 29, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Hu, H.; Kshitiz; Ahmed, I.; Levchenko, A.; Semenza, G.L. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 2013, 288, 10703–10714. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.V.; Fofo, H.; Bejarano, E.; Bento, C.F.; Ramalho, J.S.; Girao, H.; Pereira, P. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy 2013, 9, 1349–1366. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Appelhoff, R.J.; Tian, Y.M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef] [PubMed]
- Di Conza, G.; Trusso Cafarello, S.; Loroch, S.; Mennerich, D.; Deschoemaeker, S.; Di Matteo, M.; Ehling, M.; Gevaert, K.; Prenen, H.; Zahedi, R.P.; et al. The mTOR and PP2A Pathways Regulate PHD2 Phosphorylation to Fine-Tune HIF1α Levels and Colorectal Cancer Cell Survival under Hypoxia. Cell Rep. 2017, 18, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K.; Yamanaka, K.; Kamura, T.; Minato, N.; Conaway, R.C.; Conaway, J.W.; Klausner, R.D.; Pause, A. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 12436–12441. [Google Scholar] [CrossRef] [PubMed]
- Chintala, S.; Najrana, T.; Toth, K.; Cao, S.; Durrani, F.A.; Pili, R.; Rustum, Y.M. Prolyl hydroxylase 2 dependent and Von-Hippel-Lindau independent degradation of Hypoxia-inducible factor 1 and 2 α by selenium in clear cell renal cell carcinoma leads to tumor growth inhibition. BMC Cancer 2012, 12, 293. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, D.; Messing, E.M.; Wu, G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 2005, 6, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Troilo, A.; Alexander, I.; Muehl, S.; Jaramillo, D.; Knobeloch, K.P.; Krek, W. HIF1α deubiquitination by USP8 is essential for ciliogenesis in normoxia. EMBO Rep. 2014, 15, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Bremm, A.; Moniz, S.; Mader, J.; Rocha, S.; Komander, D. Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep. 2014, 15, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Zeng, L.; Yeom, C.J.; Zhu, Y.; Morinibu, A.; Shinomiya, K.; Kobayashi, M.; Hirota, K.; Itasaka, S.; Yoshimura, M.; et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1α. Nat. Commun. 2015, 6, 6153. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.T.; Kuo, Y.C.; Hung, J.J.; Huang, C.H.; Chen, W.Y.; Chou, T.Y.; Chen, Y.; Chen, Y.J.; Chen, Y.J.; Cheng, W.C.; et al. K63-polyubiquitinated HAUSP deubiquitinates HIF-1α and dictates H3K56 acetylation promoting hypoxia-induced tumour progression. Nat. Commun. 2016, 7, 13644. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Cao, Y.; Li, C.Y.; Dewhirst, M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004, 5, 429–441. [Google Scholar] [CrossRef]
- Cao, Y.; Eble, J.M.; Moon, E.; Yuan, H.; Weitzel, D.H.; Landon, C.D.; Nien, C.Y.; Hanna, G.; Rich, J.N.; Provenzale, J.M.; et al. Tumor cells upregulate normoxic HIF-1α in response to doxorubicin. Cancer Res. 2013, 73, 6230–6242. [Google Scholar] [CrossRef] [PubMed]
- Magierowski, M.; Magierowska, K.; Surmiak, M.; Hubalewska-Mazgaj, M.; Kwiecien, S.; Wallace, J.L.; Brzozowski, T. The effect of hydrogen sulfide-releasing naproxen (ATB-346) versus naproxen on formation of stress-induced gastric lesions, the regulation of systemic inflammation, hypoxia and alterations in gastric microcirculation. J. Physiol. Pharmacol. 2017, 68, 749–756. [Google Scholar] [PubMed]
- Magierowski, M.; Magierowska, K.; Hubalewska-Mazgaj, M.; Adamski, J.; Bakalarz, D.; Sliwowski, Z.; Pajdo, R.; Kwiecien, S.; Brzozowski, T. Interaction between endogenous carbon monoxide and hydrogen sulfide in the mechanism of gastroprotection against acute aspirin-induced gastric damage. Pharmacol. Res. 2016, 114, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Isaacs, J.S.; Lee, S.; Trepel, J.; Neckers, L. IL-1β-mediated up-regulation of HIF-1α via an NFkappaB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB J. 2003, 17, 2115–2117. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.A.D.; Andrade, D.R.J. HIF-1α and infectious diseases: A new frontier for the development of new therapies. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e92. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.D.; Kim, W.; Jeong, J.W.; Park, J.W.; Kim, J.E. AK-1, a SIRT2 inhibitor, destabilizes HIF-1α and diminishes its transcriptional activity during hypoxia. Cancer Lett. 2016, 373, 138–145. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyo, J.; Ryu, J.; Kim, W.; Choi, J.-S.; Jeong, J.-W.; Kim, J.-E. The Protein Phosphatase PPM1G Destabilizes HIF-1α Expression. Int. J. Mol. Sci. 2018, 19, 2297. https://doi.org/10.3390/ijms19082297
Pyo J, Ryu J, Kim W, Choi J-S, Jeong J-W, Kim J-E. The Protein Phosphatase PPM1G Destabilizes HIF-1α Expression. International Journal of Molecular Sciences. 2018; 19(8):2297. https://doi.org/10.3390/ijms19082297
Chicago/Turabian StylePyo, Jaehyuk, Jaewook Ryu, Wootae Kim, Jae-Sun Choi, Joo-Won Jeong, and Ja-Eun Kim. 2018. "The Protein Phosphatase PPM1G Destabilizes HIF-1α Expression" International Journal of Molecular Sciences 19, no. 8: 2297. https://doi.org/10.3390/ijms19082297
APA StylePyo, J., Ryu, J., Kim, W., Choi, J.-S., Jeong, J.-W., & Kim, J.-E. (2018). The Protein Phosphatase PPM1G Destabilizes HIF-1α Expression. International Journal of Molecular Sciences, 19(8), 2297. https://doi.org/10.3390/ijms19082297





