Antibacterial Mechanism of Gloverin2 from Silkworm, Bombyx mori
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis of BmGlv2
2.2. Prokaryotic Expression and Thermal Stability Analysis of BmGlv2
2.3. Antibacterial Activity of BmGlv2
2.4. Binding Efficiency of BmGlv2 to E. coli JM109
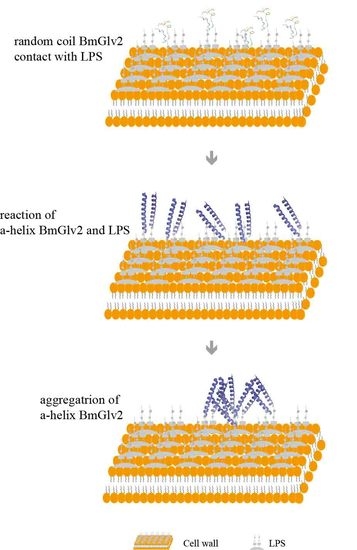
2.5. Interaction between BmGlv2 and LPS
3. Discussion
4. Materials and Methods
4.1. Sequence Alignment
4.2. Cloning of BmGlv2
4.3. Expression and Purification of BmGlv2
4.4. Circular Dichroism Spectroscopy
4.5. Antibacterial Activity Assay
4.6. Microbial Binding Assay
4.7. LPS Bingding Assay
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Mussabekova, A.; Daeffler, L.; Imler, J.L. Innate and intrinsic antiviral immunity in Drosophila. Cell. Mol. Life Sci. 2017, 74, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
- Randolph, G.J. Editorial overview: Innate immunity: The finely tuned STING of innate immunity. Curr. Opin. Immunol. 2018, 50, v–vii. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, K.; Rus, F.; Vriesema-Magnuson, C.; Erturk-Hasdemir, D.; Paquette, N.; Silverman, N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008, 4, e1000120. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [PubMed]
- Makarova, O.; Rodriguez-Rojas, A.; Eravci, M.; Weise, C.; Dobson, A.; Johnston, P.; Rolff, J. Antimicrobial defence and persistent infection in insects revisited. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Matsuki, H.; Furukawa, S.; Sagisaka, A.; Kotani, E.; Mori, H.; Yamakawa, M. Identification and functional analysis of Relish homologs in the silkworm, Bombyx mori. Biochim. Biophys. Acta 2007, 1769, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Axen, A.; Carlsson, A.; Engstrom, A.; Bennich, H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur. J. Biochem. 1997, 247, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Deng, X.J.; Yang, W.Y.; Zhou, C.Z.; Cao, Y.; Yu, X.Q. Gloverins of the silkworm Bombyx mori: Structural and binding properties and activities. Insect Biochem. Mol. Biol. 2013, 43, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Habel, D.A.; Biglang-awa, I.M.; Dulce, A.; Luu, D.D.; Garcia, P.; Weers, P.M.; Haas-Stapleton, E.J. Inactivation of the budded virus of Autographa californica M nucleopolyhedrovirus by gloverin. J. Invertebr. Pathol. 2012, 110, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, A.; Liu, G.; Kang, D.; Berzins, K.; Steiner, H. Trichoplusia ni gloverin, an inducible immune gene encoding an antibacterial insect protein. Insect Biochem. Mol. Biol. 2002, 32, 795–801. [Google Scholar] [CrossRef]
- Lu, D.; Geng, T.; Hou, C.; Huang, Y.; Qin, G.; Guo, X. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Mackintosh, J.A.; Gooley, A.A.; Karuso, P.H.; Beattie, A.J.; Jardine, D.R.; Veal, D.A. A gloverin-like antibacterial protein is synthesized in Helicoverpa armigera following bacterial challenge. Dev. Comp. Immunol. 1998, 22, 387–399. [Google Scholar] [CrossRef]
- Xu, X.X.; Jin, F.L.; Wang, Y.S.; Freed, S.; Hu, Q.B.; Ren, S.X. Molecular cloning and characterization of gloverin from the diamondback moth, Plutella xylostella L. and its interaction with bacterial membrane. World J. Microbiol. Biotechnol. 2015, 31, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Kim, Y. RNA interference of an antimicrobial peptide, gloverin, of the beet armyworm, Spodoptera exigua, enhances susceptibility to Bacillus thuringiensis. J. Invertebr. Pathol. 2011, 108, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Zhong, X.; Yi, H.Y.; Yu, X.Q. Manduca sexta gloverin binds microbial components and is active against bacteria and fungi. Dev. Comp. Immunol. 2012, 38, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, Y.; Furukawa, S.; Tanaka, H.; Yamakawa, M. Expression of antimicrobial peptide genes encoding Enbocin and Gloverin isoforms in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2007, 71, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, S.; Katsuma, S.; Daimon, T.; Isono, R.; Omuro, N.; Mita, K.; Shimada, T. Functional analysis of four Gloverin-like genes in the silkworm, Bombyx mori. Arch. Insect Biochem. Physiol. 2008, 67, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Geng, T.; Hou, C.; Qin, G.; Gao, K.; Guo, X. Expression profiling of Bombyx mori gloverin2 gene and its synergistic antifungal effect with cecropin A against Beauveria bassiana. Gene 2017, 600, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Lu, Z. Immune responses to bacterial and fungal infections in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2018, 83, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Malmsten, M. Interactions of Antimicrobial Peptides with Bacterial Membranes and Membrane Components. Curr. Top. Med. Chem. 2016, 16, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, W.; Zhang, Y.; Zhang, X.; Zhao, P.; Xia, Q. Identification and Characterization of Novel Chitin-Binding Proteins from the Larval Cuticle of Silkworm, Bombyx mori. J. Proteome Res. 2016, 15, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Lee, D.G.; Jang, S.H.; Woo, E.R.; Jeong, H.G.; Choi, C.H.; Hahm, K.S. A Leu-Lys-rich antimicrobial peptide: Activity and mechanism. Biochim. Biophys. Acta 2003, 1645, 172–182. [Google Scholar] [CrossRef]
- Rittig, M.G.; Kaufmann, A.; Robins, A.; Shaw, B.; Sprenger, H.; Gemsa, D.; Foulongne, V.; Rouot, B.; Dornand, J. Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes. J. Leukoc. Biol. 2003, 74, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, K.S.; Kim, B.Y.; Choi, Y.S.; Yoon, H.J.; Jia, J.; Jin, B.R. Anti-fibrinolytic and anti-microbial activities of a serine protease inhibitor from honeybee (Apis cerana) venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017, 201, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Liang, J.; Zhan, Z.; Xiang, Z.; He, N. Identification of the chitin-binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2010, 40, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Zelezetsky, I.; Tossi, A. Alpha-helical antimicrobial peptides—Using a sequence template to guide structure-activity relationship studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Ebenhan, T.; Gheysens, O.; Kruger, H.G.; Zeevaart, J.R.; Sathekge, M.M. Antimicrobial peptides: Their role as infection-selective tracers for molecular imaging. BioMed Res. Int. 2014, 2014, 867381. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, P.; Johnson, W.J. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 1987, 167, 76–85. [Google Scholar] [CrossRef]
- Hennessey, J.J.; Johnson, W.J. Information content in the circular dichroism of proteins. Biochemistry 1981, 20, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.K.; Miskimins, W.K. Metformin and phenethyl isothiocyanate combined treatment in vitro is cytotoxic to ovarian cancer cultures. J. Ovarian Res. 2012, 5, 19. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Guo, P.; Wang, Z.; Liu, H.; Zhang, Y.; Jiang, S.; Han, W.; Xia, Q.; Zhao, P. Antibacterial Mechanism of Gloverin2 from Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 2275. https://doi.org/10.3390/ijms19082275
Wang Q, Guo P, Wang Z, Liu H, Zhang Y, Jiang S, Han W, Xia Q, Zhao P. Antibacterial Mechanism of Gloverin2 from Silkworm, Bombyx mori. International Journal of Molecular Sciences. 2018; 19(8):2275. https://doi.org/10.3390/ijms19082275
Chicago/Turabian StyleWang, Qian, Pengchao Guo, Zhan Wang, Huawei Liu, Yunshi Zhang, Shan Jiang, Wenzhe Han, Qingyou Xia, and Ping Zhao. 2018. "Antibacterial Mechanism of Gloverin2 from Silkworm, Bombyx mori" International Journal of Molecular Sciences 19, no. 8: 2275. https://doi.org/10.3390/ijms19082275