In Vitro Comparison of 2D-Cell Culture and 3D-Cell Sheets of Scleraxis-Programmed Bone Marrow Derived Mesenchymal Stem Cells to Primary Tendon Stem/Progenitor Cells for Tendon Repair
Abstract
:1. Introduction
2. Results
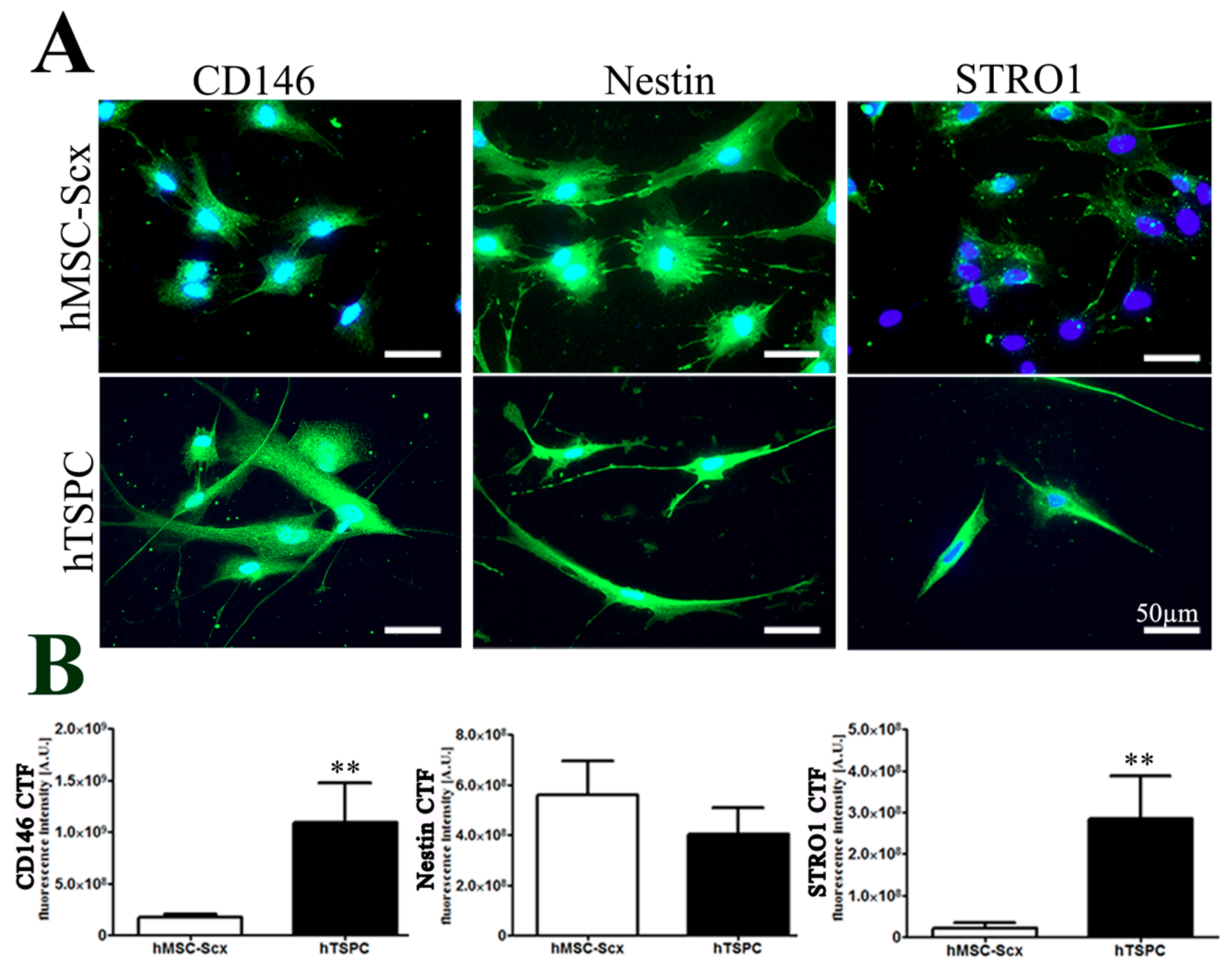
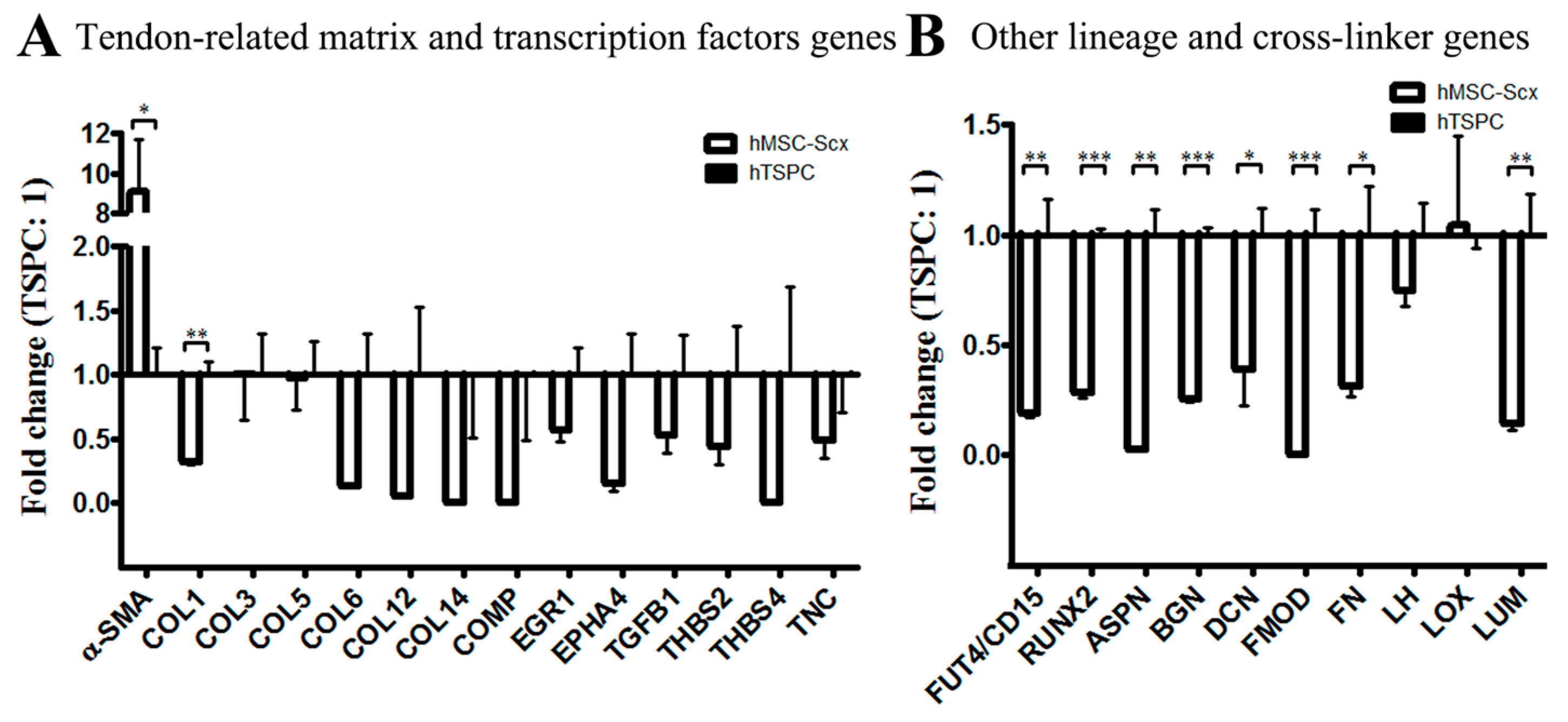
2.1. Comparison of Stem Cell Characteristic
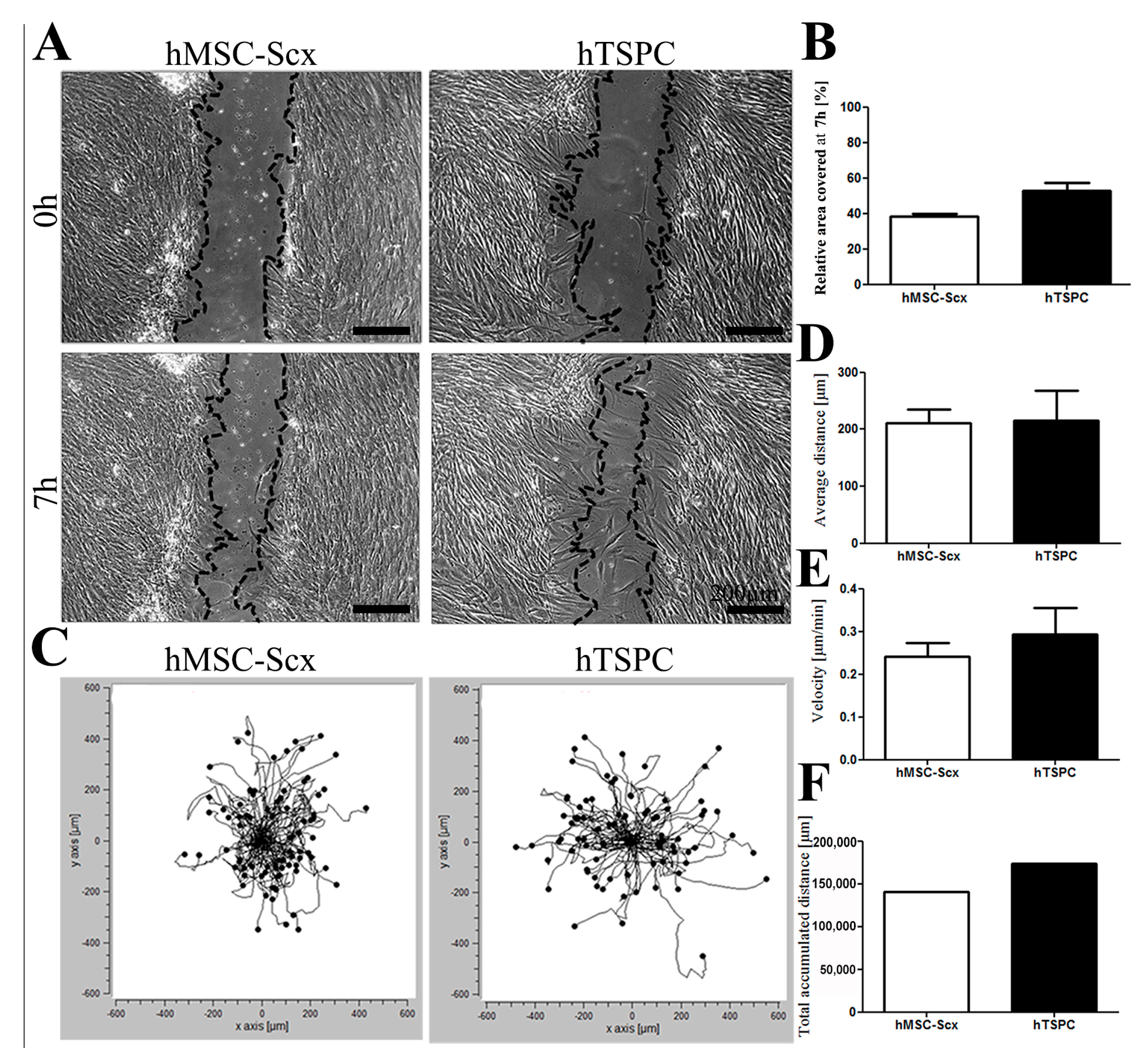
2.2. In Vitro Wound Repair Potential of the HMSC-Scx Cells Versus HTSPCs
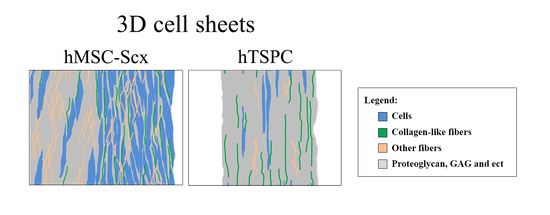
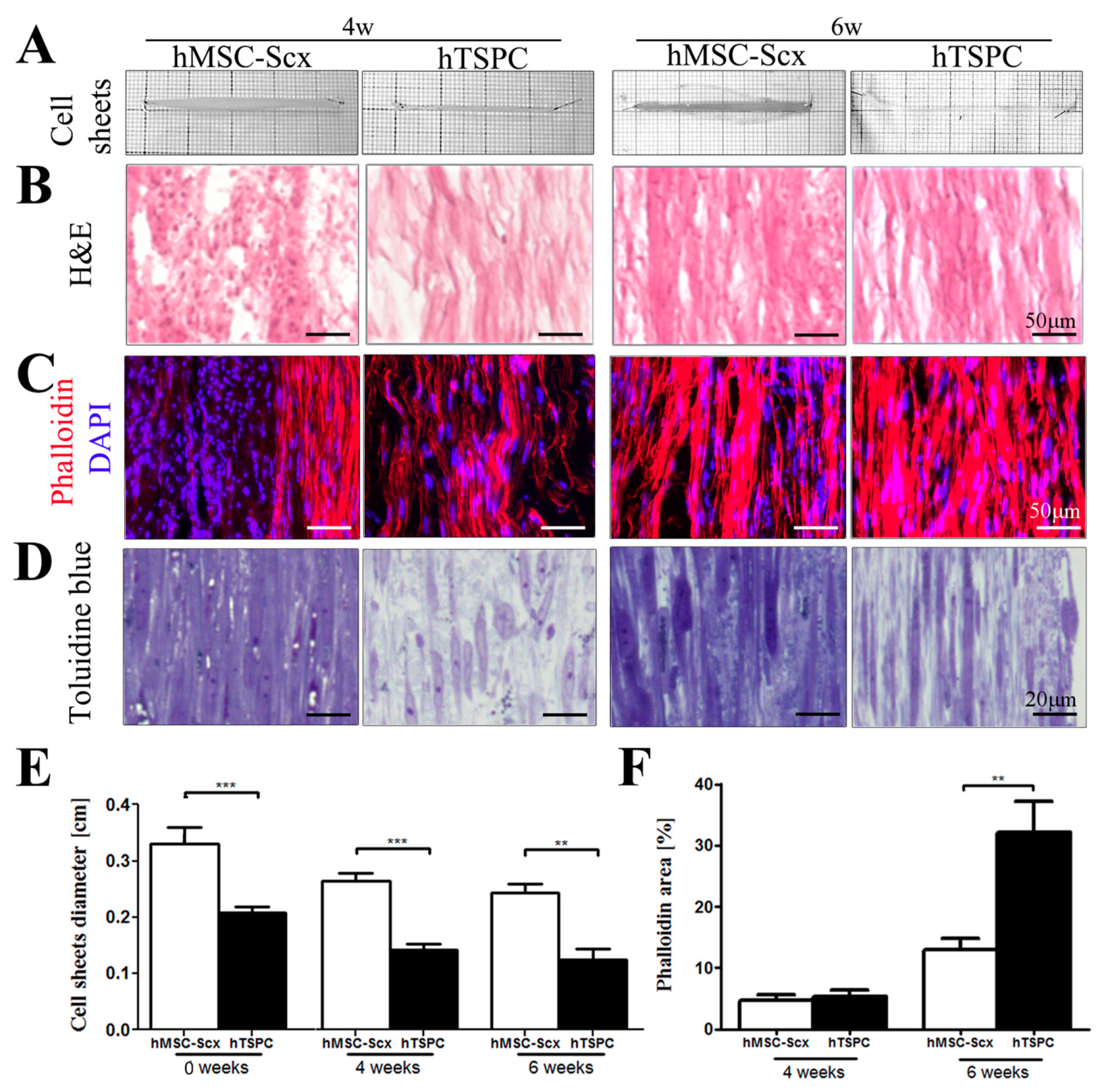
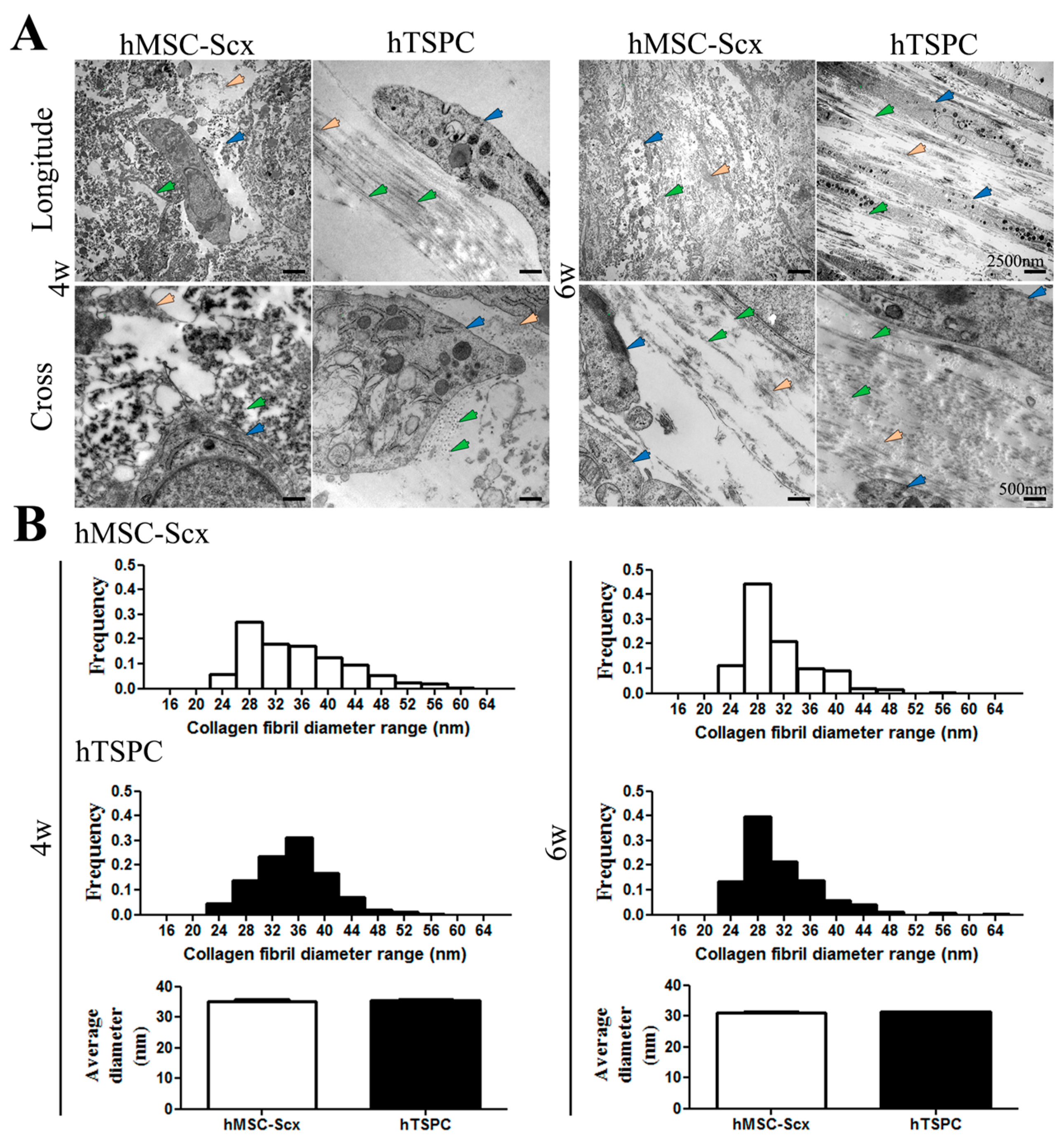
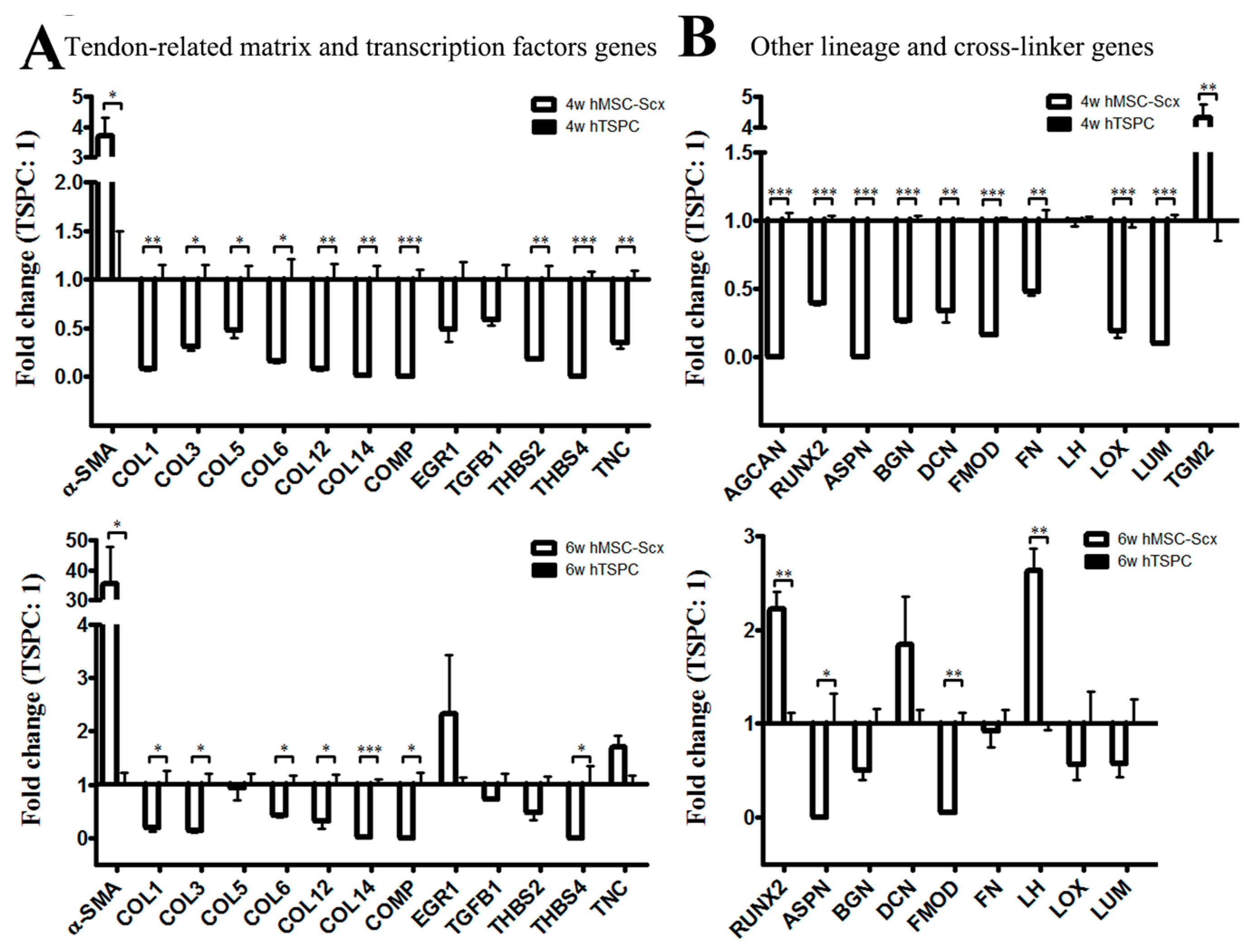
2.3. Qualitative and Quantitate Examination of Three-Dimensional hMSC-Scx and hTSPC Sheets
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunocytochemistry
4.3. PCR
4.4. Cell Migration
4.5. Cell Sheets
4.6. Cell Sheet Histology
4.7. Transmission Electron Microscopy
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, G.; Rothrauff, B.B.; Tuan, R.S. Tendon and ligament regeneration and repair: Clinical relevance and developmental paradigm. Birth Defects Res. Part C Embryo Today 2013, 99, 203–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, M.; Angele, P.; Jarvinen, T.A.H.; Docheva, D. Rescue plan for Achilles: Therapeutics steering the fate and functions of stem cells in tendon wound healing. Adv. Drug Deliv. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Yin, H.; Nerlich, M.; Pfeifer, C.G.; Docheva, D. Boosting tendon repair: Interplay of cells, growth factors and scaffold-free and gel-based carriers. J. Exp. Orthop. 2018, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Docheva, D.; Muller, S.A.; Majewski, M.; Evans, C.H. Biologics for tendon repair. Adv. Drug Deliv. Rev. 2015, 84, 222–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, Y.; Ehirchiou, D.; Kilts, T.M.; Inkson, C.A.; Embree, M.C.; Sonoyama, W.; Li, L.; Leet, A.I.; Seo, B.M.; Zhang, L.; et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 2007, 13, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.; Popov, C.; Klotz, B.; Alberton, P.; Prall, W.C.; Haasters, F.; Muller-Deubert, S.; Ebert, R.; Klein-Hitpass, L.; Jakob, F.; et al. Uncovering the cellular and molecular changes in tendon stem/progenitor cells attributed to tendon aging and degeneration. Aging Cell 2013, 12, 988–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, C.; Burggraf, M.; Kreja, L.; Ignatius, A.; Schieker, M.; Docheva, D. Mechanical stimulation of human tendon stem/progenitor cells results in upregulation of matrix proteins, integrins and MMPs, and activation of p38 and ERK1/2 kinases. BMC Mol. Biol. 2015, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Pietschmann, M.F.; Frankewycz, B.; Schmitz, P.; Docheva, D.; Sievers, B.; Jansson, V.; Schieker, M.; Muller, P.E. Comparison of tenocytes and mesenchymal stem cells seeded on biodegradable scaffolds in a full-size tendon defect model. J. Mater. Sci. Mater. Med. 2013, 24, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.F.; Alberton, P.; Loffredo-Verde, E.; Volkmer, E.; Pietschmann, M.; Muller, P.E.; Schieker, M.; Docheva, D. Periodontal ligament cells as alternative source for cell-based therapy of tendon injuries: In vivo study of full-size Achilles tendon defect in a rat model. Eur. Cells Mater. 2016, 32, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Guerquin, M.J.; Charvet, B.; Nourissat, G.; Havis, E.; Ronsin, O.; Bonnin, M.A.; Ruggiu, M.; Olivera-Martinez, I.; Robert, N.; Lu, Y.; et al. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Investig. 2013, 123, 3564–3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberton, P.; Popov, C.; Pragert, M.; Kohler, J.; Shukunami, C.; Schieker, M.; Docheva, D. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev. 2012, 21, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Gulotta, L.V.; Kovacevic, D.; Packer, J.D.; Deng, X.H.; Rodeo, S.A. Bone marrow-derived mesenchymal stem cells transduced with scleraxis improve rotator cuff healing in a rat model. Am. J. Sports Med. 2011, 39, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, C.; Zhu, S.; Lu, P.; Zhu, T.; Gong, X.; Zhang, Z.; Hu, J.; Yin, Z.; Heng, B.C.; et al. Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFbeta signaling pathway. Stem Cells 2015, 33, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.F.; Alberton, P.; Loffredo-Verde, E.; Volkmer, E.; Pietschmann, M.; Muller, P.; Schieker, M.; Docheva, D. Scaffold-free Scleraxis-programmed tendon progenitors aid in significantly enhanced repair of full-size Achilles tendon rupture. Nanomedicine 2016, 11, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, I.; Wang, J.H.; Iwasaki, K.; Shimizu, T.; Okano, T. The effect of tendon stem/progenitor cell (TSC) sheet on the early tendon healing in a rat Achilles tendon injury model. Acta Biomater. 2016, 42, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Mifune, Y.; Inui, A.; Sakata, R.; Muto, T.; Takase, F.; Ueda, Y.; Kataoka, T.; Kokubu, T.; Kuroda, R.; et al. Rotator cuff repair using cell sheets derived from human rotator cuff in a rat model. J. Orthop. Res. 2017, 35, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, S.; Zhang, C.; Lu, P.; Hu, J.; Yin, Z.; Ma, Y.; Chen, X.; OuYang, H. Crucial transcription factors in tendon development and differentiation: Their potential for tendon regeneration. Cell Tissue Res. 2014, 356, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.A.; Heyworth, C.M.; Dexter, T.M. How do stem cells decide what to do? Ciba Found. Symp. 1997, 204, 3–14. [Google Scholar] [PubMed]
- Davey, R.E.; Zandstra, P.W. Signal processing underlying extrinsic control of stem cell fate. Curr. Opin. Hematol. 2004, 11, 95–101. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, A.; Schaffer, D.V. The biology and engineering of stem-cell control. Biotechnol. Appl. Biochem. 2004, 40, 5–16. [Google Scholar] [PubMed]
- de Lucas, B.; Perez, L.M.; Galvez, B.G. Importance and regulation of adult stem cell migration. J. Cell. Mol. Med. 2018, 22, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Sohni, A.; Verfaillie, C.M. Mesenchymal stem cells migration homing and tracking. Stem Cells Int. 2013, 2013, 130763. [Google Scholar] [CrossRef] [PubMed]
- Gelberman, R.H.; Linderman, S.W.; Jayaram, R.; Dikina, A.D.; Sakiyama-Elbert, S.; Alsberg, E.; Thomopoulos, S.; Shen, H. Combined Administration of ASCs and BMP-12 Promotes an M2 Macrophage Phenotype and Enhances Tendon Healing. Clin. Orthop. Relat. Res. 2017, 475, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Garvin, J.; Qi, J.; Maloney, M.; Banes, A.J. Novel system for engineering bioartificial tendons and application of mechanical load. Tissue Eng. 2003, 9, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Riehl, B.D.; Park, J.H.; Kwon, I.K.; Lim, J.Y. Mechanical stretching for tissue engineering: Two-dimensional and three-dimensional constructs. Tissue Eng. Part B Rev. 2012, 18, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Lejard, V.; Brideau, G.; Blais, F.; Salingcarnboriboon, R.; Wagner, G.; Roehrl, M.H.; Noda, M.; Duprez, D.; Houillier, P.; Rossert, J. Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 2007, 282, 17665–17675. [Google Scholar] [CrossRef] [PubMed]
- Espira, L.; Lamoureux, L.; Jones, S.C.; Gerard, R.D.; Dixon, I.M.; Czubryt, M.P. The basic helix-loop-helix transcription factor scleraxis regulates fibroblast collagen synthesis. J. Mol. Cell. Cardiol. 2009, 47, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, R.A.; Czubryt, M.P. Synergistic roles of scleraxis and Smads in the regulation of collagen 1alpha2 gene expression. Biochim. Biophys. Acta 2012, 1823, 1936–1944. [Google Scholar] [CrossRef] [PubMed]
- Blitz, E.; Sharir, A.; Akiyama, H.; Zelzer, E. Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 2013, 140, 2680–2690. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Takimoto, A.; Akiyama, H.; Kist, R.; Scherer, G.; Nakamura, T.; Hiraki, Y.; Shukunami, C. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 2013, 140, 2280–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asou, Y.; Nifuji, A.; Tsuji, K.; Shinomiya, K.; Olson, E.N.; Koopman, P.; Noda, M. Coordinated expression of scleraxis and Sox9 genes during embryonic development of tendons and cartilage. J. Orthop. Res. 2002, 20, 827–833. [Google Scholar] [CrossRef]
- Subramanian, A.; Schilling, T.F. Tendon development and musculoskeletal assembly: Emerging roles for the extracellular matrix. Development 2015, 142, 4191–4204. [Google Scholar] [CrossRef] [PubMed]
- Im, G.I. Nonviral gene transfer strategies to promote bone regeneration. J. Biomed. Mater. Res. A 2013, 101, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, S.A. In vivo nonviral delivery factors to enhance bone repair. Clin. Orthop. Relat. Res. 2000, 379, S113–S119. [Google Scholar] [CrossRef]
- Partridge, K.A.; Oreffo, R.O. Gene delivery in bone tissue engineering: Progress and prospects using viral and nonviral strategies. Tissue Eng. 2004, 10, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Breidahl, W.; Mackie, K.E.; Lin, Z.; Qin, A.; Chen, J.; Zheng, M.H. Autologous tenocyte injection for the treatment of severe, chronic resistant lateral epicondylitis: A pilot study. Am. J. Sports Med. 2013, 41, 2925–2932. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, X.H.; Yin, Z.; Zou, X.H.; Wang, L.L.; Hu, H.; Cao, T.; Zheng, M.; Ouyang, H.W. Stepwise differentiation of human embryonic stem cells promotes tendon regeneration by secreting fetal tendon matrix and differentiation factors. Stem Cells 2009, 27, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Dex, S.; Alberton, P.; Willkomm, L.; Sollradl, T.; Bago, S.; Milz, S.; Shakibaei, M.; Ignatius, A.; Bloch, W.; Clausen-Schaumann, H.; et al. Tenomodulin is Required for Tendon Endurance Running and Collagen I Fibril Adaptation to Mechanical Load. EBioMedicine 2017, 20, 240–254. [Google Scholar] [CrossRef] [PubMed]
| Tenogenic-Related Genes | 2D Cell Culture | 3D Cell Sheets | ||||
|---|---|---|---|---|---|---|
| 4w | 6w | |||||
| Cells (S: hMSC-Scx; T: hTSPC) | S | T | S | T | S | T |
| Alpha smooth muscle actin (α-SMA) | ||||||
| Cartilage oligomeric matrix protein (COMP) | ||||||
| Collagen, type I, alpha 1 (COL1A1) | ||||||
| Collagen, type III, alpha 1 (COL3A1) | ||||||
| Collagen, type V, alpha 1 (COL5A1) | ||||||
| Collagen, type VI, alpha 1 (COL6A1) | ||||||
| Collagen, type XII, alpha 1 (COL12A1) | ||||||
| Collagen, type XIV, alpha 1 (COL14A1) | ||||||
| Collagen, type XV, alpha 1 (COL15A1) | ||||||
| Early growth response 1 (EGR-1) | ||||||
| Early growth response 2 (EGR-2) | ||||||
| Ephrin type-A receptor 4 (EPHA4) | ||||||
| Eyes absent homolog 1 (EYA1) | ||||||
| Eyes absent homolog 2 (EYA2) | ||||||
| Mohawk homeobox (MKX) | ||||||
| Proteoglycan 4 (PRG4) | ||||||
| Scleraxis homolog A (SCXA) | ||||||
| SIX homeobox1 (SIX1) | ||||||
| SIX homeobox2 (SIX2) | ||||||
| Tenascin C (TNC) | ||||||
| Tenomodulin (Tnmd) | ||||||
| Thrombospondin 2 (THBS2) | ||||||
| Thrombospondin 4 (THBS4) | ||||||
| Transforming growth factor beta 1 (TGF-β1) | ||||||
| Lineage & Cross-Linking Genes | 2D Cell Culture | 3D Cell Sheets | |||||
|---|---|---|---|---|---|---|---|
| 4w | 6w | ||||||
| Cells (S: hMSC-Scx; T: hTSPC) | S | T | S | T | S | T | |
| Adipogenic- related | Adipocyte Protein 2 (AP2) | ||||||
| Lipoprotein lipase (LPL) | |||||||
| Peroxisome proliferator-activated receptor gamma (PPARG) | |||||||
| Chondrogenic- related | Aggrecan (AGCAN) | ||||||
| Collagen, type II, alpha 1 (COL2A1) | |||||||
| SRY (sex-determining region Y)-box 9 (SOX9) | |||||||
| Embryonic- related | Fucosyltransferase 4 (FUT4/CD15) | ||||||
| Nanog homeobox (NANOG) | |||||||
| Octamer-binding transcription factor 4 (Oct4, Oct3) | |||||||
| Muscle- related | Desmin (DES) | ||||||
| Myogenic differentiation 1 (MYOD1) | |||||||
| Myogenin (MYOG) | |||||||
| Osteogenic- related | Integrin binding sialoprotein (IBSP) | ||||||
| Osterix (OSX) | |||||||
| Runt-related transcription factor 2 (RUNX2) | |||||||
| Collagen cross linker | Asporin (ASPN) | ||||||
| Biglycan (BGN) | |||||||
| Decorin (DCN) | |||||||
| Fibromodulin (FMOD) | |||||||
| Fibronectin (FN) | |||||||
| Lumican (LUM) | |||||||
| Lysyl hydroxylases (LH) | |||||||
| Lysyl oxidase (LOX) | |||||||
| Transglutaminase 2 (TGM2) | |||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-F.; Yan, Z.; Schumann, R.G.; Milz, S.; Pfeifer, C.G.; Schieker, M.; Docheva, D. In Vitro Comparison of 2D-Cell Culture and 3D-Cell Sheets of Scleraxis-Programmed Bone Marrow Derived Mesenchymal Stem Cells to Primary Tendon Stem/Progenitor Cells for Tendon Repair. Int. J. Mol. Sci. 2018, 19, 2272. https://doi.org/10.3390/ijms19082272
Hsieh C-F, Yan Z, Schumann RG, Milz S, Pfeifer CG, Schieker M, Docheva D. In Vitro Comparison of 2D-Cell Culture and 3D-Cell Sheets of Scleraxis-Programmed Bone Marrow Derived Mesenchymal Stem Cells to Primary Tendon Stem/Progenitor Cells for Tendon Repair. International Journal of Molecular Sciences. 2018; 19(8):2272. https://doi.org/10.3390/ijms19082272
Chicago/Turabian StyleHsieh, Chi-Fen, Zexing Yan, Ricarda G. Schumann, Stefan Milz, Christian G. Pfeifer, Matthias Schieker, and Denitsa Docheva. 2018. "In Vitro Comparison of 2D-Cell Culture and 3D-Cell Sheets of Scleraxis-Programmed Bone Marrow Derived Mesenchymal Stem Cells to Primary Tendon Stem/Progenitor Cells for Tendon Repair" International Journal of Molecular Sciences 19, no. 8: 2272. https://doi.org/10.3390/ijms19082272