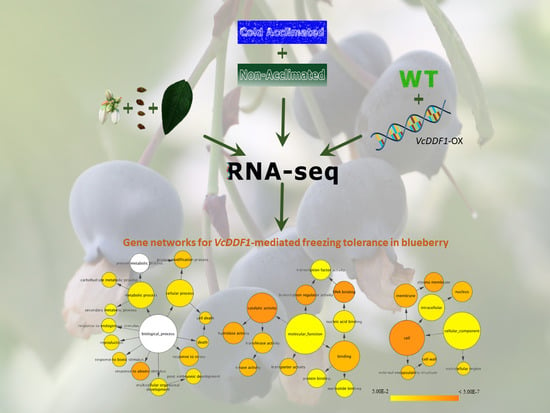
The Cold-Regulated Genes of Blueberry and Their Response to Overexpression of VcDDF1 in Several Tissues
Abstract
:1. Introduction
2. Results
2.1. Orthologues of Arabidopsis Cold-Regulated Genes in Blueberry
2.2. VcCORs and Freezing Tolerance
2.3. Expression of VcCORs in Legacy-VcDDF1-OX Plants
2.4. Blueberry CBF/DREB1 Genes
2.5. Validation of the Expression of VcDDF1
3. Discussion
3.1. VcCOR Genes in Blueberry
3.2. VcDDF1 and Freezing Tolerance
4. Method
4.1. Plant Materials
4.2. RNA Preparation, Sequencing, and de Novo Transcriptome Assembly
4.3. Differential Expression Analysis and Transcriptome Annotation
4.4. Expressed Sequence Tags (ESTs) in Blueberry
4.5. Identification of the VcCORs
4.6. QRT-PCR Analysis
4.7. Gene Network Construction
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Pearce, R.S. Plant freezing and damage. Ann. Bot. 2001, 87, 417–424. [Google Scholar] [CrossRef]
- Li, C.Y.; Puhakainen, T.; Welling, A.; Vihera-Aarnio, A.; Ernstsen, A.; Junttila, O.; Heino, P.; Pavla, E.T. Cold acclimation in silver birch (Betula pendula). Development of freezing tolerance in different tissues and climatic ecotypes. Physiol. Plant. 2002, 116, 478–488. [Google Scholar] [CrossRef]
- Strimbeck, G.R.; Schaberg, P.G.; Fossdal, C.G.; Schroder, W.P.; Kjellsen, T.D. Extreme low temperature tolerance in woody plants. Front. Plant Sci. 2015, 6, 884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larcher, W. Physiological Plant Ecology: Ecophysiology and Stress Physiology of Functional Groups, 4th ed.; Springer: Berlin, Germany; New York, NY, USA, 2003; p. xx. 513p. [Google Scholar]
- Kreyling, J.; Schmid, S.; Aas, G. Cold tolerance of tree species is related to the climate of their native ranges. J. Biogeogr. 2015, 42, 156–166. [Google Scholar] [CrossRef]
- Chuine, I.C.; Bonhomme, M.; Legave, J.-M.; De Cortázar-atauri, I.; Charrier, G.; Lacointe, A.; Améglio, T. Can phenological models predict tree phenology accurately in the future? The unrevealed hurdle of endodormancy break. Glob. Chang. Biol. 2016, 22, 3444–3460. [Google Scholar] [CrossRef] [PubMed]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Parmesan, C.; Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.J.; Brennan, R.M.; Jones, H.G. Declining chilling and its impact on temperate perennial crops. Environ. Exp. Bot. 2013, 91, 48–62. [Google Scholar] [CrossRef]
- Luedeling, E.; Girvetz, E.H.; Semenov, M.A.; Brown, P.H. Climate change affects winter chill for temperate fruit and nut trees. PLoS ONE 2011, 6, e20155. [Google Scholar] [CrossRef] [PubMed]
- Melke, A. The Physiology of Chilling Temperature Requirements for Dormancy Release and Bud-break in Temperate Fruit Trees Grown at Mild Winter Tropical Climate. J. Plant Stud. 2015, 4, 110–156. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Huang, J.; Sun, T.; Wang, X.; Zhu, C.; Ai, Y.; Gu, H. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant. Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Berdeja, M.; Nicolas, P.; Kappel, C.; Dai, Z.W.; Hilbert, G.; Peccoux, A.; Lafontaine, M.; Ollat, N.; Gomes, E.; Delrot, S. Water limitation and rootstock genotype interact to alter grape berry metabolism through transcriptome reprogramming. Hortic. Res. 2015, 2, 15012. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F.; Gilmour, S.J.; Stockinger, E.J.; Jaglo-Ottosen, K.R.; Zarka, D.G. Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol. Plant. 2001, 112, 171–175. [Google Scholar] [CrossRef]
- Miller, A.K.; Galiba, G.; Dubcovsky, J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-A(m)2 in Triticum monococcum. Mol. Genet. Genom. 2006, 275, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Carvallo, M.A.; Pino, M.T.; Jeknic, Z.; Zou, C.; Doherty, C.J.; Shiu, S.H.; Chen, T.H.H.; Thomashow, M.F. A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3807–3819. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- El Kayal, W.; Navarro, M.; Marque, G.; Keller, G.; Marque, C.; Teulieres, C. Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. J. Exp. Bot. 2006, 57, 2455–2469. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Ayax, C.; Martinez, Y.; Laur, J.; El Kayal, W.; Marque, C.; Teulieres, C. Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol. J. 2011, 9, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015, 6, 85. [Google Scholar] [CrossRef] [PubMed]
- Tillett, R.L.; Wheatley, M.D.; Tattersall, E.A.; Schlauch, K.A.; Cramer, G.R.; Cushman, J.C. The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol. J. 2012, 10, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Oakenfull, R.J.; Baxter, R.; Knight, M.R. A C-repeat binding factor transcriptional activator (CBF/DREB1) from European bilberry (Vaccinium myrtillus) induces freezing tolerance when expressed in Arabidopsis thaliana. PLoS ONE 2013, 8, e54119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.M.; Dong, J.; Hu, Y.L.; Lin, Z.P.; Xu, X.F.; Han, Z.H. Improved Cold-resistant Performance in Transgenic Grape (Vitis vinifera L.) Overexpressing Cold-inducible Transcription Factors AtDREB1b. Hortscience 2009, 44, 35–39. [Google Scholar]
- Owens, C.L.; Thomashow, M.F.; Hancock, J.F.; Iezzoni, A.F. CBF1 orthologs in sour cherry and strawberry and the heterologous expression of CBF1 in strawberry. J. Am. Soc. Hortic. Sci. 2002, 127, 489–494. [Google Scholar]
- USDA-NASS. Noncitrus Fruits and Nuts—2016 Summary. June 2017. Available online: http://usda.mannlib.cornell.edu/usda/current/NoncFruiNu/NoncFruiNu-06-27-2017.pdf (accessed on 7 March 2018).
- Longstroth, M.; Hanson, E. The Michigan Blueberry Industry. Michigan State University Extension. 2012, p. 5. Available online: http://msue.anr.msu.edu/uploads/files/The_Michigan_Blueberry_Industry_2012_MSUE_online.pdf (accessed on 7 March 2018).
- Rowland, L.J.; Ogden, E.L.; Ehlenfeldt, M.K.; Arora, R. Cold Tolerance of Blueberry Genotypes Throughout the Dormant Period from Acclimation to Deacclimation. Hortscience 2008, 43, 1970–1974. [Google Scholar]
- Walworth, A.E.; Rowland, L.J.; Polashock, J.J.; Hancock, J.F.; Song, G.Q. Overexpression of a blueberry-derived CBF gene enhances cold tolerance in a southern highbush blueberry cultivar. Mol. Breed. 2012, 30, 1313–1323. [Google Scholar] [CrossRef]
- Song, G.Q.; Gao, X. Transcriptomic changes reveal gene networks responding to the overexpression of a blueberry DWARF AND DELAYED FLOWERING 1 gene in transgenic blueberry plants. BMC Plant Biol. 2017, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Main, D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Polashock, J.J.; Arora, R.; Peng, Y.; Naik, D.; Rowland, L.J. Functional Identification of a C-repeat Binding Factor Transcriptional Activator from Blueberry Associated with Cold Acclimation and Freezing Tolerance. J. Am. Soc. Hortic. Sci. 2010, 135, 40–48. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Walworth, A.E.; Chai, B.; Song, G.Q. Transcript Profile of Flowering Regulatory Genes in VcFT-Overexpressing Blueberry Plants. PLoS ONE 2016, 11, e0156993. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]




| Source of Transcripts | Transcriptome Comparison | Percentage (Number) of AtCORs that Have DE Blueberry Orthologues) | Percentage (Number) of Unique Genes out of the Annotation of the DE VcCORs ** (Annotated by Trinotate) | Total Number of AtCORs that Have (DE) Blueberry Orthologues) | Total Number of Unique Genes out of the Annotation of the (DE) VcCORs (Annotated by Trinotate) |
|---|---|---|---|---|---|
| Blueberry transcriptome reference: reftrinity | 2181 | 5326 | |||
| Bluecrop leaf (MID5) * | 511 | 685 | |||
| Bluecrop bud (MID10: 0 CU) * | 687 | 996 | |||
| Bluecrop bud (MID4: 397 CU) * | 620 | 874 | |||
| Bluecrop bud (MID1: 789 CU) * | 610 | 873 | |||
| Bluecrop bud (MID2: 1333 CU) * | 553 | 802 | |||
| Bluecrop leaf and bud (MID5, MID10, MID4, MID1, and MID2) * | 1169 | 1960 | |||
| Legacy | Leaf versus flower | 49.2% (1074/2181) | 58.7% (3126/5326) | (1074) | (3126) |
| Flower versus non-acclimated bud | 52.9% (1154/2181) | 69.1% (3678/5326) | (1154) | (3678) | |
| Leaf versus non-acclimated bud | 50.8% (1108/2181) | 61.8% (3293/5326) | (1108) | (3293) | |
| Flower versus chilled bud | 51.9% (1131/2181) | 64.5% (3434/5326) | (1131) | (3434) | |
| Chilled bud versus non-acclimated bud | 43.7% (953/2181) | 49.5% (2639/5326) | (953) | (2639) | |
| Legacy-VcDDF1-OX | Leaf versus flower | 49.9% (1089/2181) | 59.7% (3178/5326) | 1089 | (3178) |
| Flower versus non-acclimated bud | 52.7% (1150/2181) | 67.7% (3606/5326) | 1150 | (3606) | |
| Leaf versus non-acclimated bud | 53.0% (1156/2181) | 66.5% (3542/5326) | 1156 | (3542) | |
| Legacy and Legacy-VcDDF1-OX | Legacy-VcDDF1-OX leaf versus Legacy leaf | 12.9% (282/2181) | 7.4% (396/5326) | 282 | (396) |
| Legacy-VcDDF1-OX flower versus Legacy flower | 11.4% (248/2181) | 6.9% (365/5326) | 248 | (365) | |
| Non-acclimated Legacy-VcDDF1-OX bud versus non-acclimated Legacy bud | 17.8% (389/2181) | 12.1% (646/5326) | 389 | (646) |
| Transcript | Arabidopsis1 | Transcription Factors | E-Value | Annotation | Log2Fold Change (Legacy-VcDDF1-OX/Legacy) | Log2Fold Change for Legacy | Log2Fold Change for Legacy-VcDDF1-OX | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | Non-Acclimated Buds | Flower | Chill Bud/Non-Acclimated Bud | Flower/Chilled Buds | Leaf/Flower | Flower/Non-Acclimated Bud | Leaf/Non-Acclimated Bud | Specificity | Leaf/Flower | Flower/Non-Acclimated Bud | Leaf/Non-Acclimated Bud | |||||
| c88132_g2_i2 | AT4G25490 | CBF1 | 2.00 × 10−21 | DRE1B_ARATH | 3.21 | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | leaf = bud = flower | 3.70 | #N/A | 3.93 |
| c75369_g2_i1 | AT4G25470 | CBF2 | 3.00 × 10−21 | ERF38_ARATH | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | leaf = bud = flower | 3.08 | #N/A | 1.76 |
| c85919_g2_i1 | AT4G25470 | CBF2, CBF1, CBF4, CBF3, DDF1 | 5.00 × 10−45 | DRE1F_ORYSJ | #N/A | #N/A | #N/A | #N/A | 4.50 | −1.88 | 2.67 | #N/A | leaf = bud < flower | #N/A | #N/A | #N/A |
| c85919_g2_i2 | AT4G25470 | CBF2, CBF1, CBF4, CBF3, DDF1 | 7.00 × 10−45 | DRE1F_ORYSJ | #N/A | #N/A | #N/A | #N/A | 3.10 | −1.87 | 2.05 | #N/A | leaf = bud < flower | #N/A | #N/A | #N/A |
| c85919_g2_i4 | AT4G25470 | CBF2, CBF1, CBF4, CBF3, DDF1 | 3.00 × 10−45 | DRE1F_ORYSJ | #N/A | #N/A | #N/A | −2.20 | 5.80 | −2.48 | 4.35 | #N/A | leaf = bud < flower | #N/A | 4.04 | 4.03 |
| c85919_g2_i5 | AT4G25470 | CBF2, CBF1, CBF4, CBF3, DDF1 | 6.00 × 10−45 | DRE1F_ORYSJ | #N/A | #N/A | #N/A | #N/A | 4.50 | −2.30 | 5.45 | #N/A | leaf = bud < flower | #N/A | 4.23 | 4.84 |
| c85919_g2_i6 | AT4G25470 | CBF2, CBF1, CBF4, CBF3, DDF1 | 3.00 × 10−45 | DRE1F_ORYSJ | #N/A | #N/A | #N/A | #N/A | 5.10 | −3.42 | 6.06 | #N/A | leaf = bud < flower | #N/A | 4.97 | 4.42 |
| c82156_g1_i1 | AT4G25470 | CBF2, DDF1 | 1.00 × 10−22 | ERF23_ARATH | #N/A | #N/A | #N/A | #N/A | −3.90 | 6.35 | −4.69 | 1.67 | flower < bud < leaf | 5.32 | −2.57 | 2.78 |
| c91057_g4_i1 | AT4G25470 | CBF2, DDF1, CBF1 | 2.00 × 10−26 | ERF43_ARATH | #N/A | #N/A | #N/A | −1.80 | −5.30 | #N/A | −7.56 | −6.37 | leaf = flower < bud | #N/A | −9.70 | −8.40 |
| c97417_g2_i1 | AT4G25470 | CBF2, DDF1, CBF1, CBF3, CBF4 | 2.00 × 10−27 | TINY_ARATH | #N/A | #N/A | −0.65 | #N/A | 3.10 | −1.24 | 2.63 | 1.37 | bud < leaf < flower | −1.57 | 2.69 | 1.13 |
| c88132_g2_i1 | AT4G25480 | CBF3, CBF2, CBF1, CBF4, DDF1 | 2.00 × 10−61 | DRE1A_ARATH | 2.47 | −1.87 | −2.96 | #N/A | 2.90 | −2.63 | #N/A | −2.06 | leaf < bud = flower | 2.78 | #N/A | 2.21 |
| c77615_g1_i1 | AT1G12610 | DDF1, CBF2 | 4.00 × 10−27 | DREB3_ARATH | #N/A | #N/A | #N/A | #N/A | −1.00 | 2.58 | −1.61 | #N/A | flower < leaf = bud | #N/A | #N/A | #N/A |
| c87707_g1_i1 | AT1G12610 | DDF1, CBF2 | 5.00 × 10−24 | DREB3_ARATH | #N/A | #N/A | #N/A | #N/A | −0.90 | 1.51 | −1.16 | #N/A | flower < leaf = bud | 1.46 | −1.20 | #N/A |
| c91057_g4_i3 | AT1G12610 | DDF1, CBF2 | 2.00 × 10−23 | DREB3_ARATH | #N/A | #N/A | #N/A | −1.00 | −7.50 | 3.96 | −8.75 | −4.81 | Flower < leaf < bud | 2.69 | −8.23 | −5.54 |
| c32575_g1_i1 | AT1G12610 | DDF1, CBF2, CFB3, CBF1, CBF4 | 2.00 × 10−42 | DRE1E_ARATH | 7.64 | 7.27 | 4.54 | −2.20 | 4.80 | −2.26 | 3.25 | #N/A | leaf = bud < flower | #N/A | #N/A | 1.31 |
| c62996_g1_i1 | AT1G12610 | DDF1, CBF2, CFB3, CBF1, CBF4 | 5.00 × 10−44 | DRE1E_ARATH | 4.61 | #N/A | 3.96 | #N/A | #N/A | #N/A | #N/A | #N/A | leaf = bud = flower | #N/A | #N/A | #N/A |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walworth, A.; Song, G.-q. The Cold-Regulated Genes of Blueberry and Their Response to Overexpression of VcDDF1 in Several Tissues. Int. J. Mol. Sci. 2018, 19, 1553. https://doi.org/10.3390/ijms19061553
Walworth A, Song G-q. The Cold-Regulated Genes of Blueberry and Their Response to Overexpression of VcDDF1 in Several Tissues. International Journal of Molecular Sciences. 2018; 19(6):1553. https://doi.org/10.3390/ijms19061553
Chicago/Turabian StyleWalworth, Aaron, and Guo-qing Song. 2018. "The Cold-Regulated Genes of Blueberry and Their Response to Overexpression of VcDDF1 in Several Tissues" International Journal of Molecular Sciences 19, no. 6: 1553. https://doi.org/10.3390/ijms19061553
APA StyleWalworth, A., & Song, G.-q. (2018). The Cold-Regulated Genes of Blueberry and Their Response to Overexpression of VcDDF1 in Several Tissues. International Journal of Molecular Sciences, 19(6), 1553. https://doi.org/10.3390/ijms19061553