Fine Mapping and Candidate Gene Prediction for White Immature Fruit Skin in Cucumber (Cucumis sativus L.)
Abstract
:1. Introduction
2. Results
2.1. Genetic Analysis of the Immature Fruit Skin Color Trait
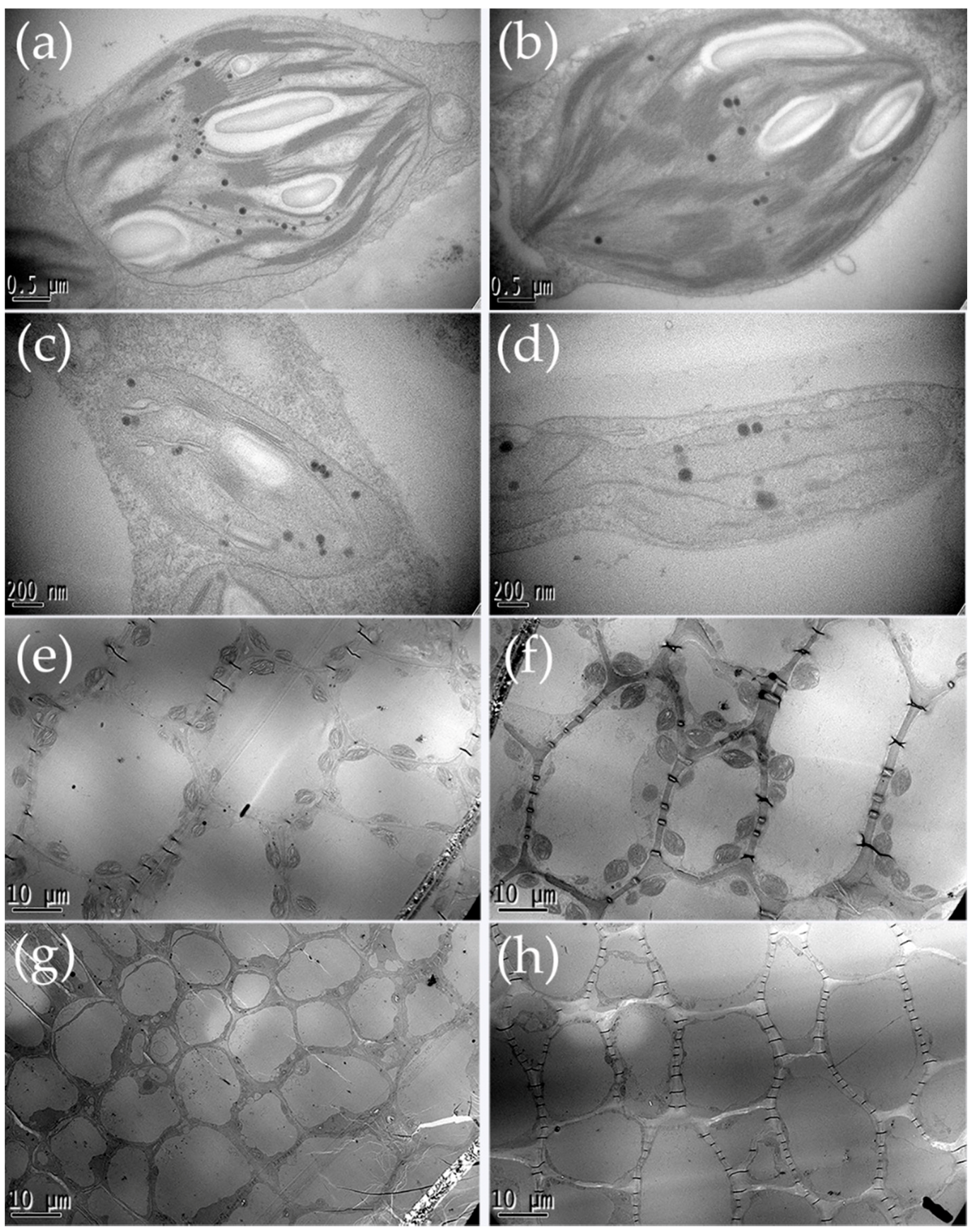
2.2. Chlorophyll Content Determination and Chloroplast Observation
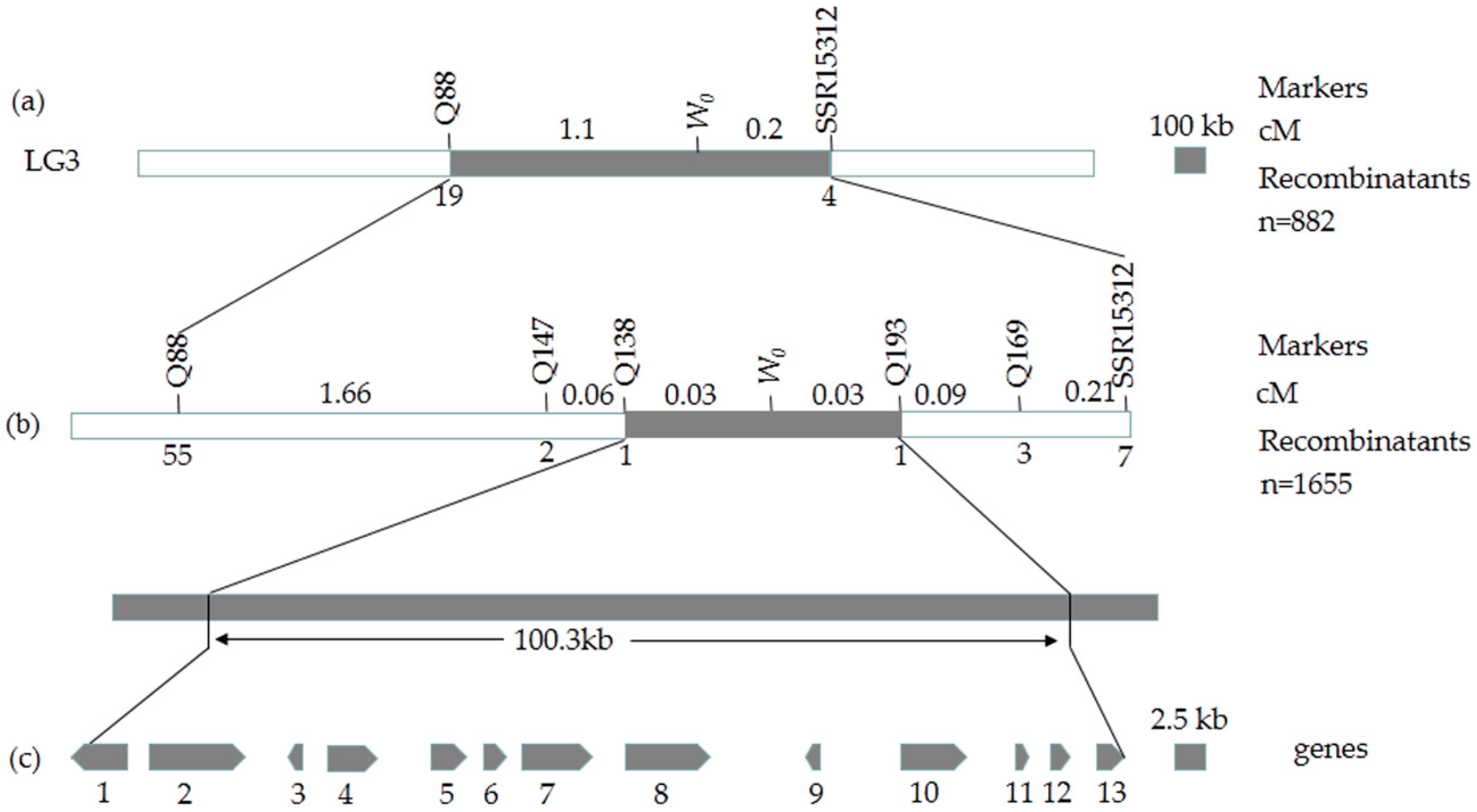
2.3. Preliminary Mapping of the White Immature Fruit Skin Color Trait
2.4. Fine Mapping of the White Immature Fruit Skin Color Trait
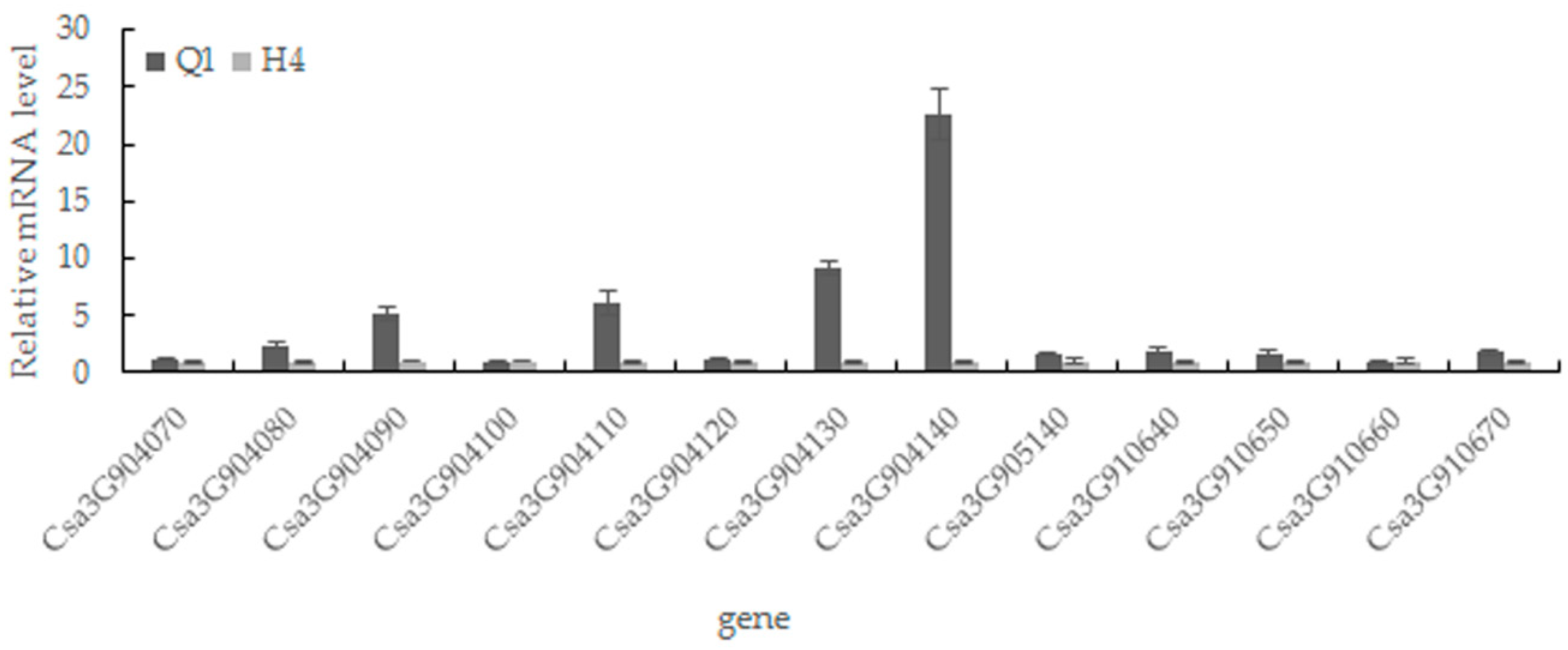
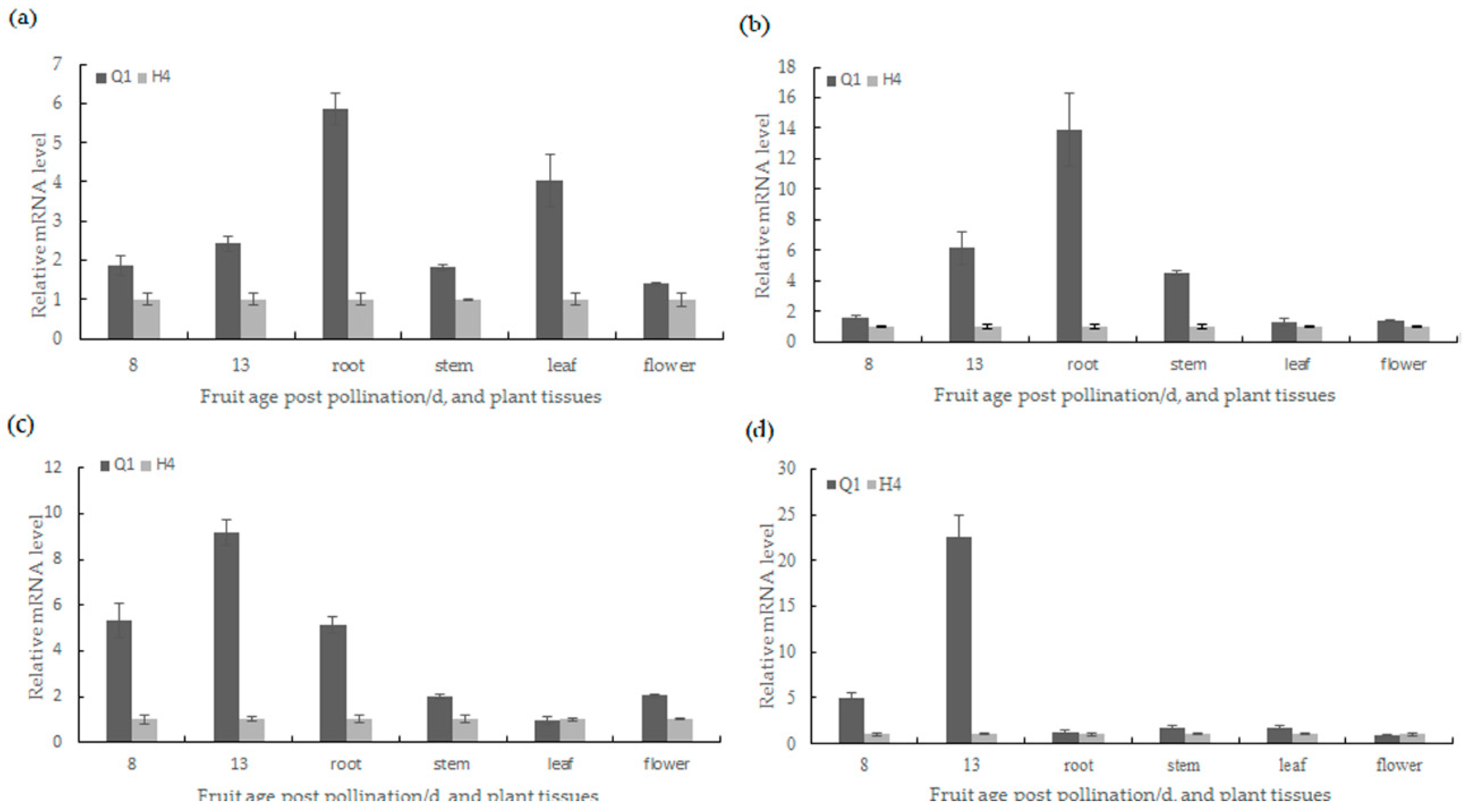
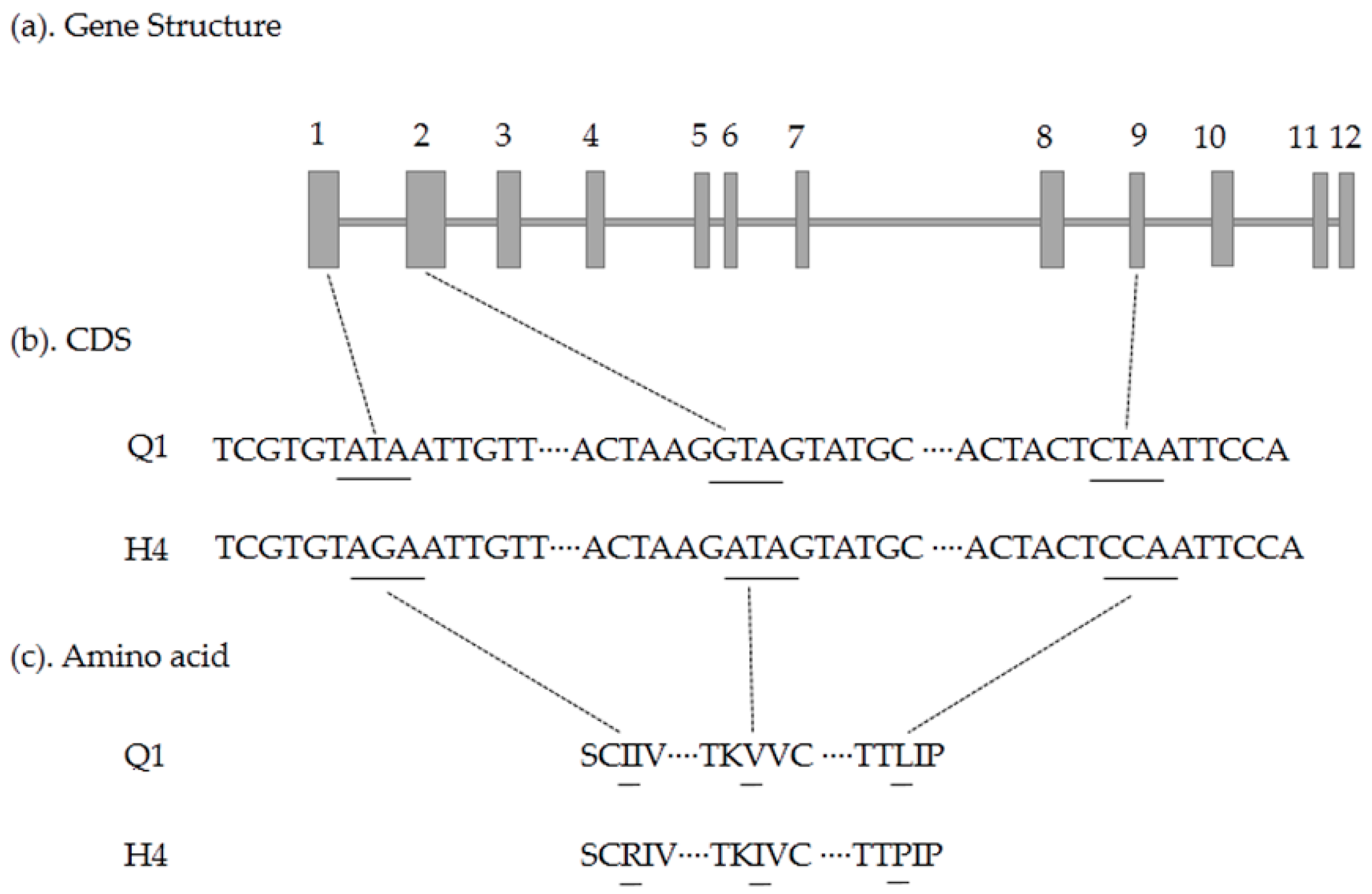
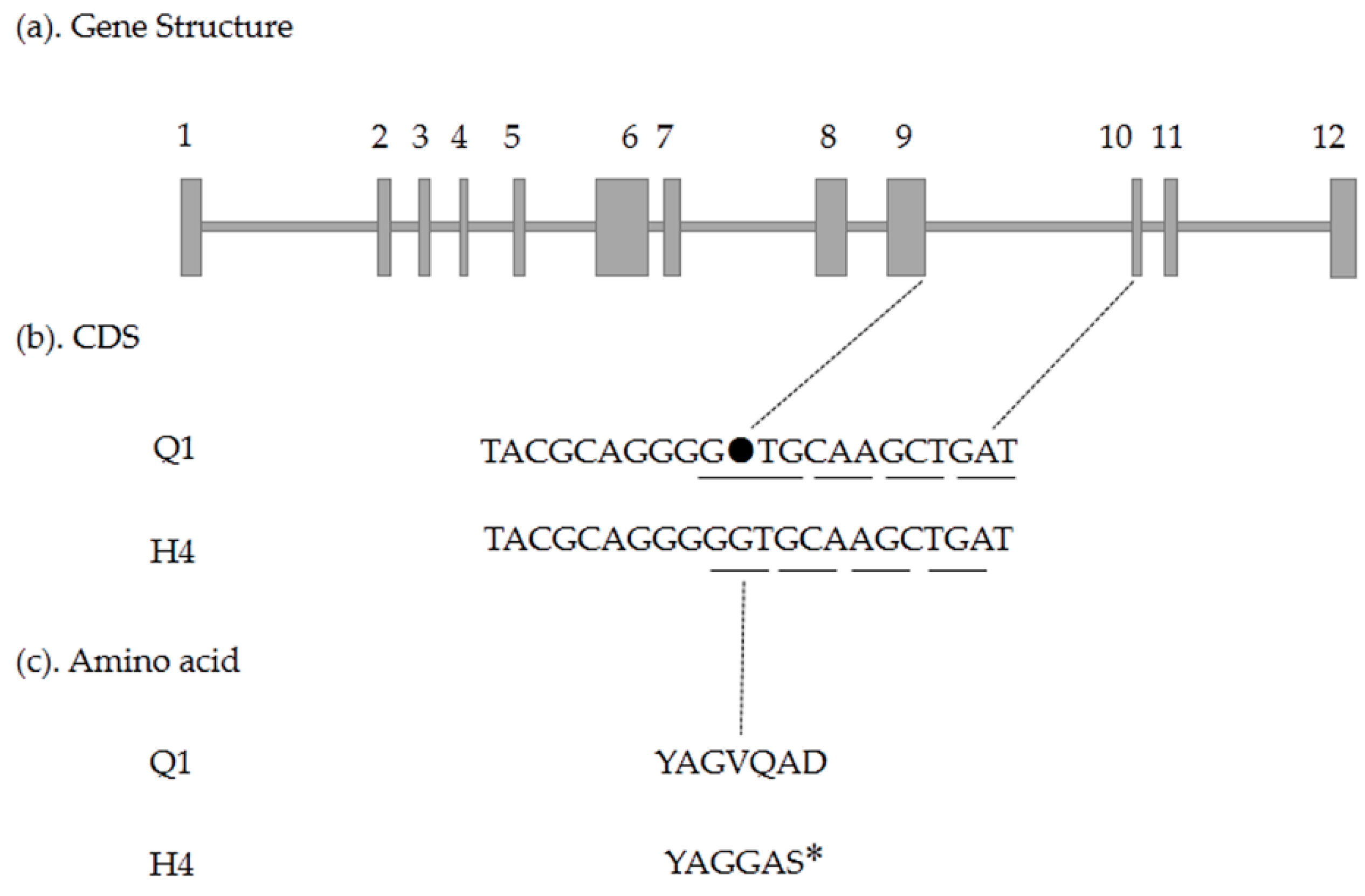
2.5. Candidate Gene Prediction and Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Genetic Mapping Population
4.2. Chlorophyll Content Determination and Chloroplast Observation
4.3. Design of SSR and CAPS Markers
4.4. Gene Prediction and Expression Analysis
- 18sRNA-F: AGAAACGGCTACCACATC
- 18sRNA-R: CCAAGGTCCAACTACGAG
4.5. Candidate Gene Cloning and Sequencing
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, X.Q.; Li, Y.; Zhang, W.W.; He, H.L.; Pan, J.S.; Cai, R. Fine mapping of the uniform immature fruit color gene u in cucumber (Cucumis sativus L.). Euphytica 2014, 196, 341–348. [Google Scholar] [CrossRef]
- Huang, H.Y.; Hsieh, C.H. Genetic research on fruit color traits of the bitter gourd (Momordica charantia L.). Hortic. J. 2017, 86, 238–243. [Google Scholar] [CrossRef]
- Jiang, B.; Liu, W.R.; Xie, D.S.; Peng, Q.W.; He, X.M.; Lin, Y.E.; Liang, Z.J. High-density genetic map construction and gene mapping of pericarp color in wax gourd using specific-locus amplified fragment (SLAF) sequencing. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.B.; Wu, X.X.; Chen, J.L. Study on heredity of fruit colors of sweet pepper. Acta Hortic. Sin. 2005, 32, 513–515. [Google Scholar]
- Wu, L.; Liu, J.Y.; Liang, Y. Inheritance on fruit color character between green and red of tomato. Acta Hortic. Sin. 2016, 43, 674–682. [Google Scholar]
- Jin, F.M.; Xue, J.; Jia, Y.H.; Liu, Z.Q. Research advance of tomato fruit color related gene. Tianjin Agric. Sci. 2006, 12, 3–6. [Google Scholar]
- Sooriyapathirana, S.S.; Khan, A.; Sebolt, A.M.; Wang, D.C.; Bushakra, J.M.; Lin-Wang, K.; Allan, A.C.; Gardiner, S.E.; Chagne, D.; Iezzoni, A.F. QTL analysis and candidate gene mapping for skin and flesh color in sweet cherry fruit (Prunus avium L.). Tree Genet. Genom. 2010, 6, 821–832. [Google Scholar] [CrossRef]
- Jin, W.M.; Wang, H.; Li, M.F.; Wang, J.; Yang, Y.; Zhang, X.M.; Yan, G.H.; Zhang, H.; Liu, J.S.; Zhang, K.C. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Wen, C.L.; Weng, Y.Q. Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor. Appl. Genet. 2013, 126, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.J.; Pan, J.S.; Cai, R.; Guan, Y.; Liu, L.Z.; Zhang, W.W.; Li, Z.; He, H.L.; Zhang, C.; Si, L.T.; et al. Genetic mapping and QTL analysis of fruit and flower related traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Euphytica 2008, 164, 473–491. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, S.P.; Wang, X.W.; Zhang, Z.H.; Li, M.; Mu, S.Q.; Cheng, Z.C.; Zhang, R.W.; Huang, S.W.; Xie, B.Y.; et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 2011, 182, 167–176. [Google Scholar] [CrossRef]
- Dong, S.Y.; Miao, H.; Zhang, S.P.; Liu, M.M.; Wang, Y.; Gu, X.F. Genetic analysis and gene mapping of white fruit skin in cucumber (Cucumis sativus L.). Acta Bot. Boreal. Occident. Sin. 2012, 11, 2177–2181. [Google Scholar]
- Wang, J.K.; Fang, X.X.; Li, X.H.; Chen, Y.; Wan, Z.J.; Xu, Y.J. Genetic study on immature fruit color of cucumber. Acta Hortic. Sin. 2013, 128, 479–486. [Google Scholar]
- Liu, H.; Jiao, J.Q.; Liang, X.J.; Liu, J.; Meng, H.W.; Chen, S.X.; Li, Y.H.; Cheng, Z.H. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, Z.H.; Liu, J.H.; Staub, J.E.; Han, Y.H.; Cheng, Z.C.; Li, X.F.; Lu, J.Y.; Miao, H.; Kang, H.X.; et al. Integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 2009, 4, e5795. [Google Scholar] [CrossRef] [PubMed]
- He, J.P.; Ke, L.P.; Hong, D.F.; Xie, Y.Z.; Wang, G.C.; Liu, P.W.; Yang, G.S. Fine mapping of a recessive genic male sterility gene (Bnms3) in rapeseed (Brassica napus) with AFLP- and Arabidopsis-derived PCR markers. Theor. Appl. Genet. 2008, 117, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Liu, M.W.; Wang, S.Q.; Deng, Q.M.; Zheng, A.P.; Liu, H.N.; Wang, L.X.; Zhu, J.; Li, P. Fine mapping of a dominant minute-grain gene, Mi3, in rice. Mol. Breed. 2012, 30, 1045–1051. [Google Scholar] [CrossRef]
- Dijkhuizen, A.; Kennard, W.C.; Havey, M.J.; Staub, J.E. RFLP variation and genetic relationships in cultivated cucumber. Euphytica 1996, 90, 79–87. [Google Scholar]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.M.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.W.; Weng, Y.Q. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Baranowskij, N.; Frohberg, C.; Prat, S.; Willmitzer, L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994, 13, 5383–5392. [Google Scholar] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Lysenko, E.A. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007, 26, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Goldschmidt-Clermont, M.; Wostrikoff, K.; Rochaix, J.-D. Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 2000, 21, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.G.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Kiba, T.; Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R.; Lichtenthaler, H. Formulae and program to determine total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Adv. Photosynth. Res. 1984, 2, 9–12. [Google Scholar]
- Zhang, W.W.; He, H.L.; Guan, Y.; Du, H.; Yuan, L.H.; Li, Z.; Yao, D.Q.; Pan, J.S.; Cai, R. Identification and mapping of molecular markers linked to the tuberculate fruit gene in the cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2010, 120, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.H.; Seemann, J.R. Extraction and purification of RNA from plant tissue enriched in polysaccharides. Methods Mol. Biol. 1998, 86, 27–32. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Population | Observation | Expected Separation Ratio | χ2 | p Value | |
|---|---|---|---|---|---|
| Green | White | ||||
| BCP1 a | 120 | 0 | 1:0 | - | - |
| BCP2 b | 65 | 55 | 1:1 | 0.68 | 0.411 |
| F2 c | 115 | 29 | 3:1 | 1.56 | 0.211 |
| Gene ID | Cucurbit Genomics Database Description |
|---|---|
| Csa3G904070 | Putative peptide/nitrate transporter; contains IPR000109 (Proton-dependent oligopeptide transporter family), IPR016196 (Major facilitator superfamily domain, general substrate transporter) |
| Csa3G904080 | Pyruvate kinase; contains IPR001697 (Pyruvate kinase) |
| Csa3G904090 | Unknown protein; contains IPR008502 (Prolamin-like domain) |
| Csa3G904100 | Ribose-phosphate pyrophosphokinase; contains IPR005946 (Ribose-phosphate diphosphokinase) |
| Csa3G904110 | Tobamovirus multiplication 2B |
| Csa3G904120 | Peroxidase; contains IPR010255 (Haem peroxidase) |
| Csa3G904130 | Tetraspanin family protein; contains IPR018499 (Tetraspanin/Peripherin) |
| Csa3G904140 | Two-component response regulator-like protein; contains IPR009057 (Homeodomain-like), IPR011006 (CheY-like superfamily) |
| Csa3G905140 | Unknown protein |
| Csa3G910640 | Unknown protein; contains IPR018996 (Inner nuclear membrane protein MAN1) |
| Csa3G910650 | Allyl alcohol dehydrogenase-like protein |
| Csa3G910660 | DnaJ homolog subfamily C member; contains IPR009057 (Homeodomain-like) |
| Csa3G910670 | Expansin L; contains IPR007117 (Expansin, cellulose-binding-like domain), IPR014733 (Barwin-like endoglucanase) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, H.-Y.; Dong, X.; Wang, J.-K.; Xia, J.-H.; Xie, F.; Zhang, Y.; Yao, X.; Xu, Y.-J.; Wan, Z.-J. Fine Mapping and Candidate Gene Prediction for White Immature Fruit Skin in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2018, 19, 1493. https://doi.org/10.3390/ijms19051493
Tang H-Y, Dong X, Wang J-K, Xia J-H, Xie F, Zhang Y, Yao X, Xu Y-J, Wan Z-J. Fine Mapping and Candidate Gene Prediction for White Immature Fruit Skin in Cucumber (Cucumis sativus L.). International Journal of Molecular Sciences. 2018; 19(5):1493. https://doi.org/10.3390/ijms19051493
Chicago/Turabian StyleTang, Hong-Yu, Xu Dong, Jian-Ke Wang, Jun-Hui Xia, Fei Xie, Yu Zhang, Xuan Yao, Yue-Jin Xu, and Zheng-Jie Wan. 2018. "Fine Mapping and Candidate Gene Prediction for White Immature Fruit Skin in Cucumber (Cucumis sativus L.)" International Journal of Molecular Sciences 19, no. 5: 1493. https://doi.org/10.3390/ijms19051493




