Allosteric Activation of GDP-Bound Ras Isoforms by Bisphenol Derivative Plasticisers
Abstract
:1. Introduction
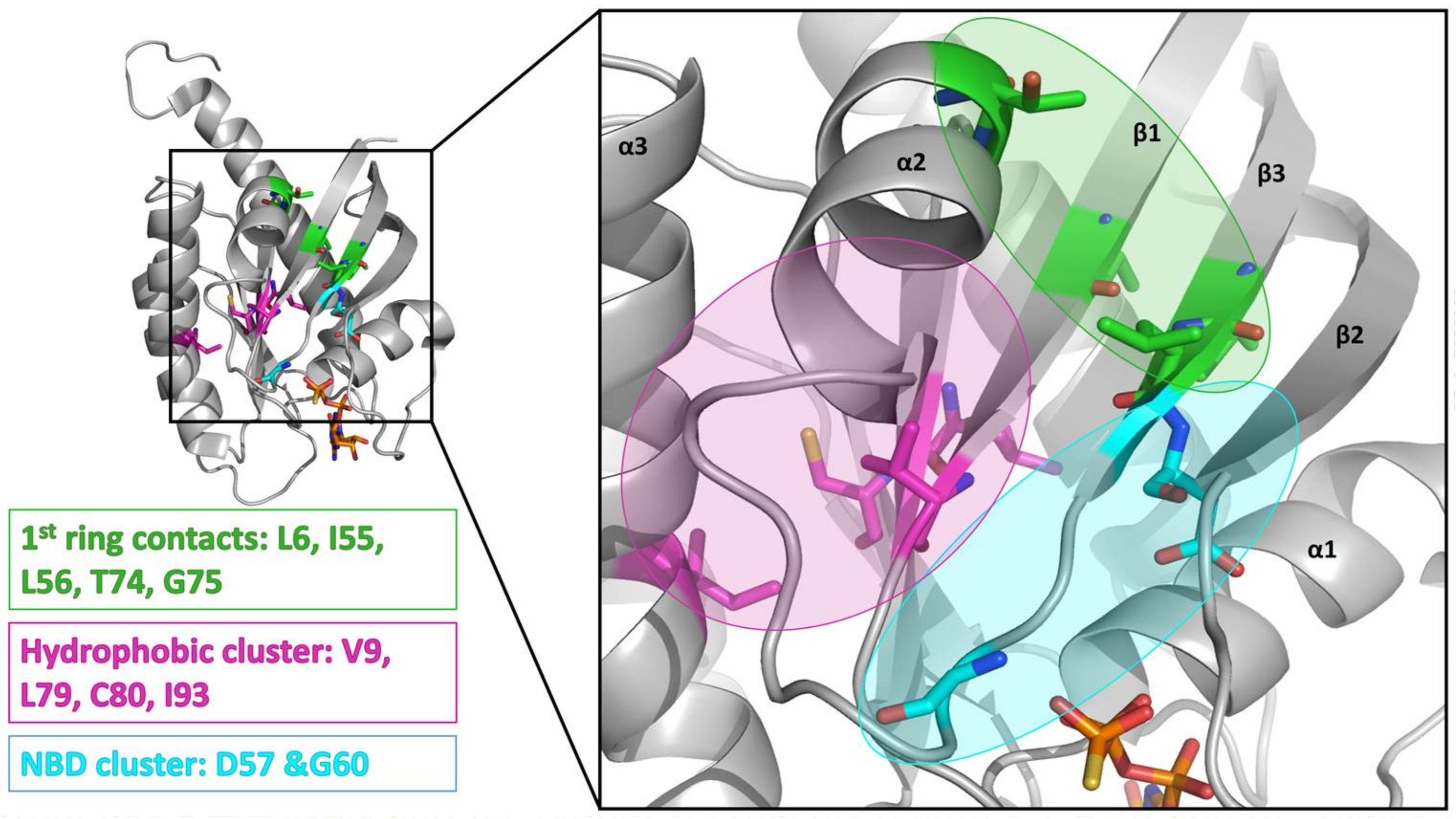
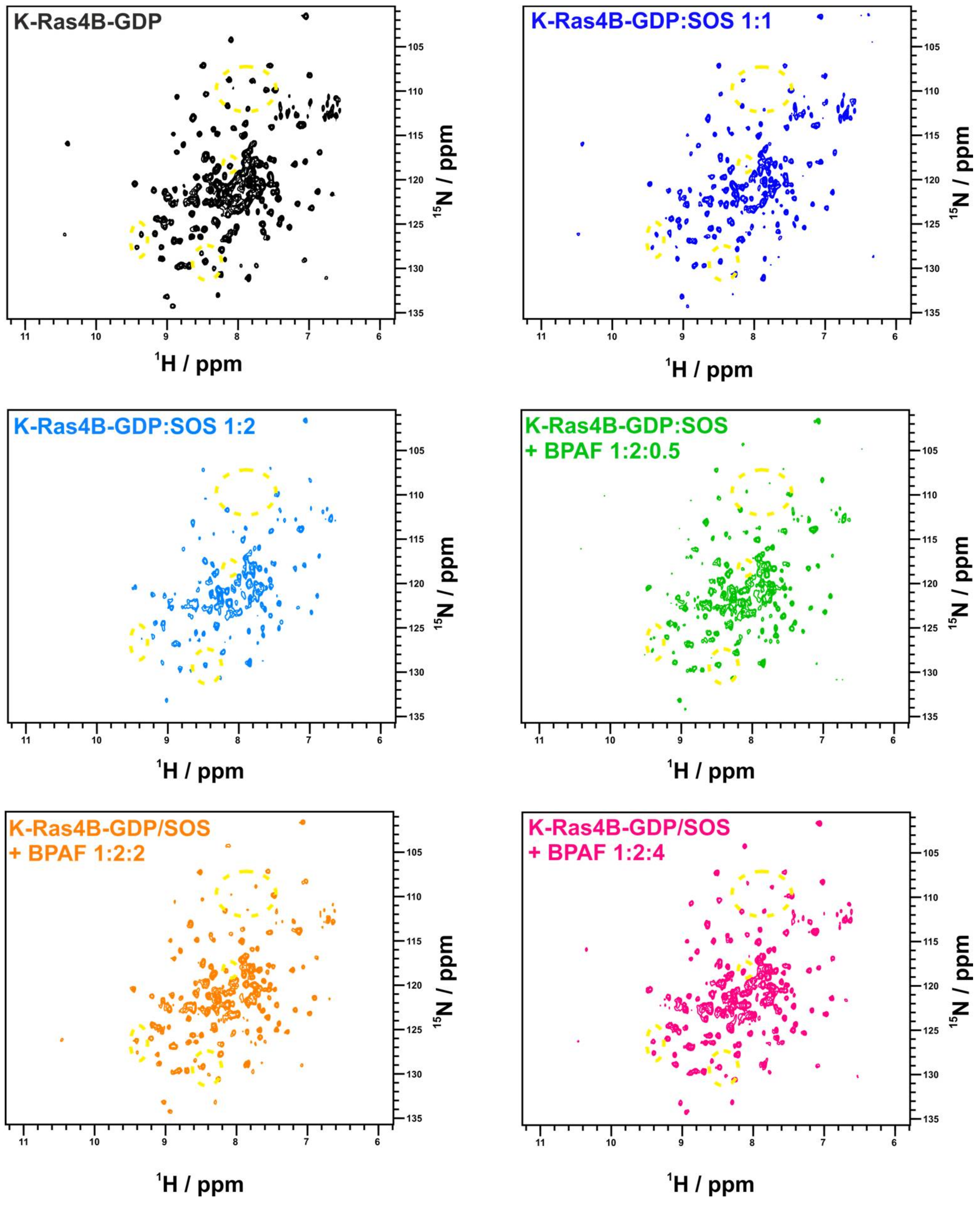
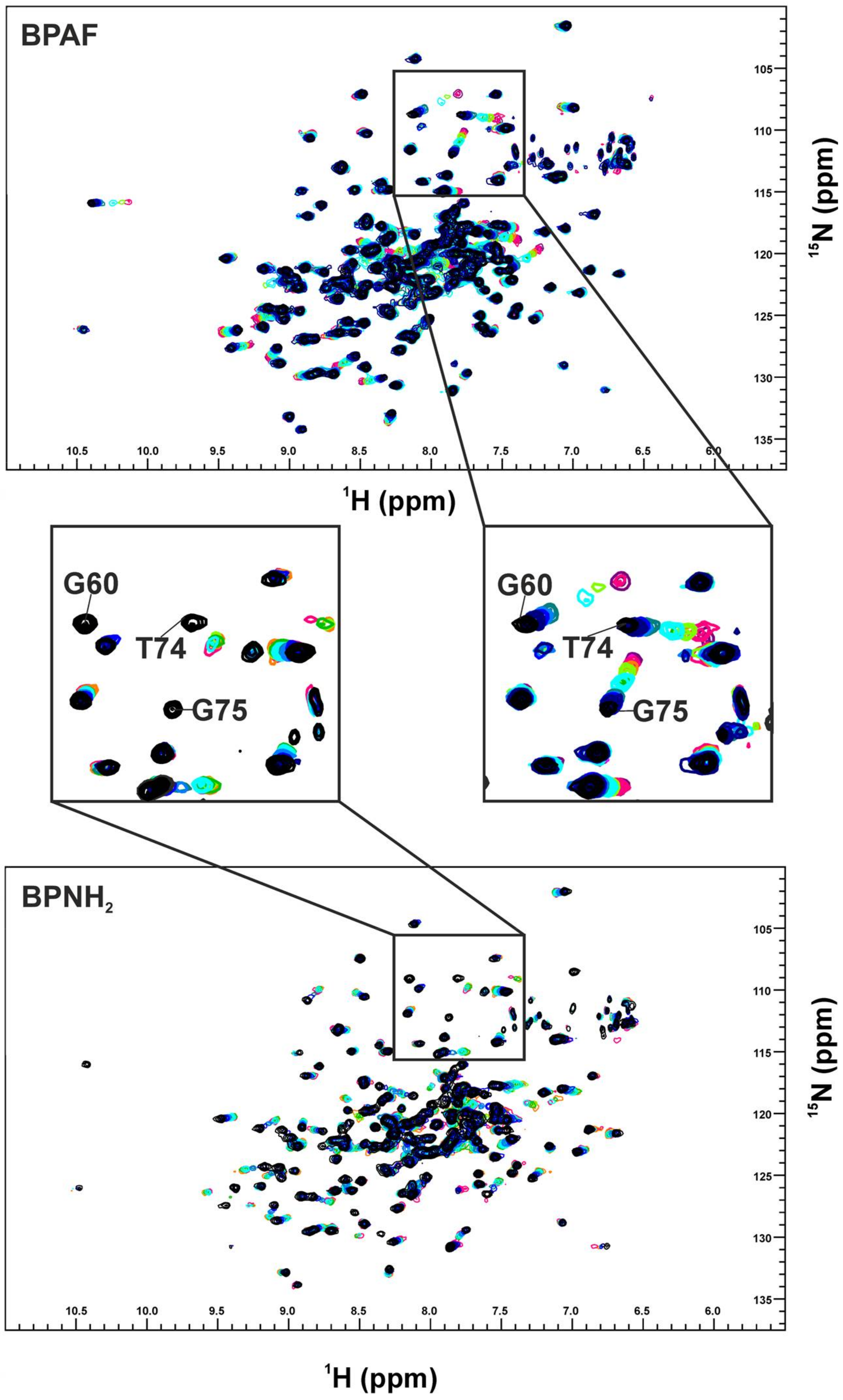
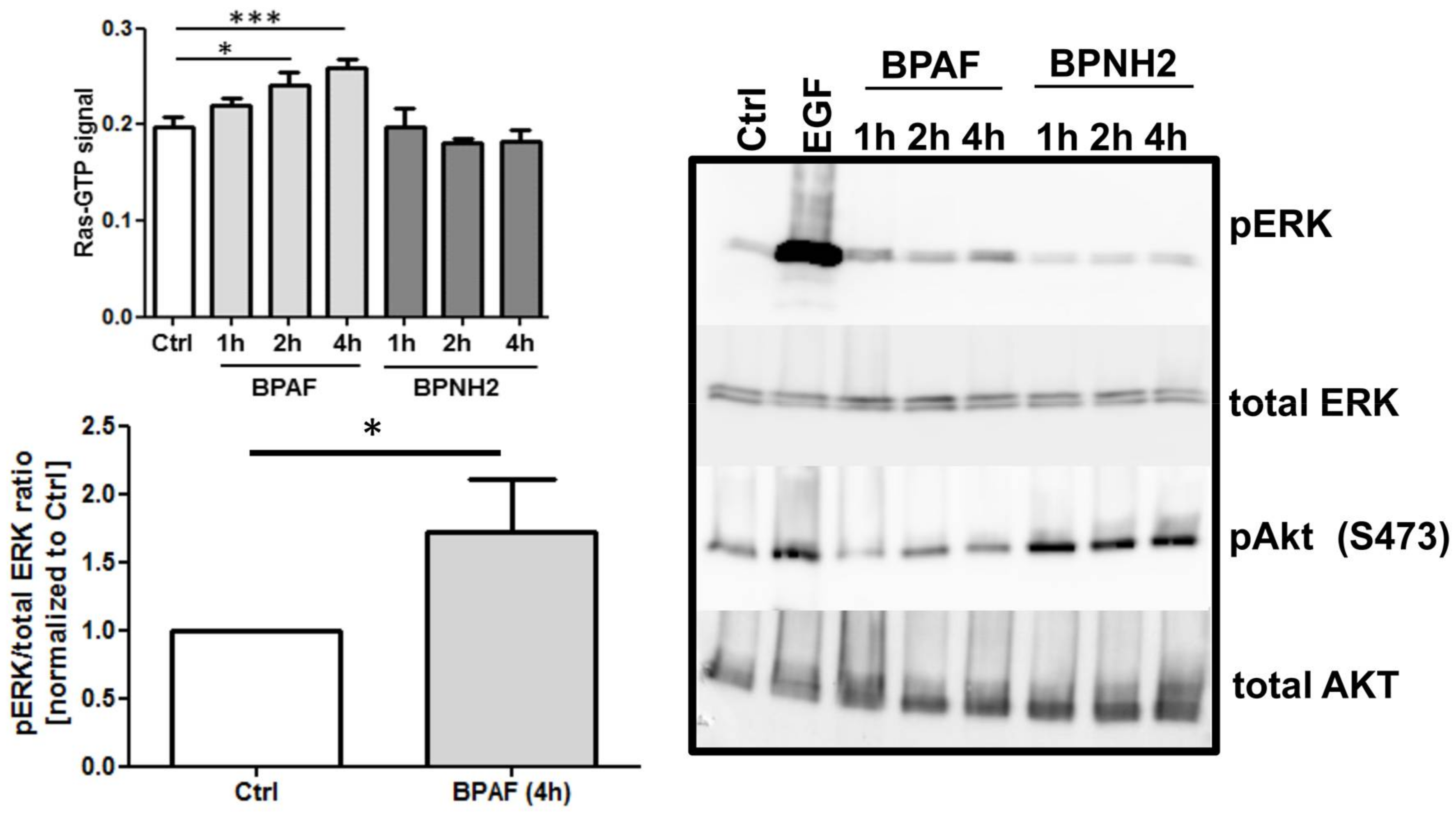
2. Results
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Low Molecular Weight Compounds and Solvents
- AFX (4-(Trifluoromethyl)phenol, CAS 402-45-9),
- Bisphenol A (BPA, 2,2-Bis(4-hydroxyphenyl)propane, CAS 80-05-7),
- Bisphenol AF (BPAF, 2,2-Bis(4-hydroxyphenyl)hexafluoropropane, CAS 1478-61-1),
- Bisphenol AP (BPAP, 4,4′-(1-Phenylethylidene)bisphenol, CAS 571-75-1),
- Bisphenol BP (BPBP, Bis(4-hydroxyphenyl)diphenylmethane, CAS 1844-01-5),
- Bisphenol C (BPC, 2,2-Bis(4-hydroxy-3-methylphenyl)propane, CAS 79-97-0),
- Bisphenol FL (BPFL, 4,4′-(9-Fluorenylidene)diphenol, CAS 3236-71-3),
- Bisphenol M (BPM, 4,4′-(1,3-Phenylenediisopropylidene)bisphenol, CAS 13595-25-0),
- Bisphenol NH2 (BPNH2, 2,2-Bis(3-amino-4-hydroxyphenyl)hexafluoropropane, CAS 83558-87-6),
- Bisphenol P (BPP, 4,4′-(1,4-Phenylenediisopropylidene)bisphenol, CAS 2167-51-3),
- Bisphenol S (BPS, 4,4′-Sulfonyldiphenol, CAS 80-09-1),
- Bisphenol Z (BPZ, 4,4′-Cyclohexylidenebisphenol, CAS 843-55-0)
- Bisphenol B (BPB, 2,2-Bis(4-hydroxyphenyl)butane, CAS 77-40-7),
- Bisphenol E (BPE, 1,1-Bis(4-hydroxyphenyl)ethane, CAS 2081 08 5),
- Bisphenol F (BPF, 4,4′-Methylenediphenol, CAS 620-92-8)
4.3. NMR Spectroscopy
4.4. Molecular Modelling
4.5. SOScat-Mediated Nucleotide Exchange Assay and GDI Assay
4.6. MTT Cytotoxicity Assay
4.7. G-LISA Ras Activation Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GDP | Guanosine Diphosphate |
| GTP | Guanosine Triphosphate |
| SOS | Son of Sevenless |
| SAR | Structure-Activity-Relationship |
| GDI | Guanine Nucleotide Dissociation |
| NMR | Nuclear Magnetic Resonance |
| CSP | Chemical Shift Perturbation |
References
- Vetter, I.R.; Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Schubbert, S.; Shannon, K.; Bollag, G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 2007, 7, 295–308. [Google Scholar] [CrossRef]
- Simon, M.A.; Bowtell, D.D.L.; Dodson, G.S.; Laverty, T.R.; Rubin, G.M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 1991, 67, 701–716. [Google Scholar] [CrossRef]
- Sharma, A.; Luke, C.T.; Dower, N.A.; Stone, J.C.; Lorenzo, P.S. RasGRP1 is essential for Ras activation by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate in epidermal keratinocytes. J. Biol. Chem. 2010, 285, 15724–15730. [Google Scholar] [CrossRef]
- Maurer, T.; Garrenton, L.S.; Oh, A.; Pitts, K.; Anderson, D.J.; Skelton, N.J. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 2012, 109, 5299–5304. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Burke, J.P.; Phan, J.; Burns, M.C.; Olejniczak, E.T.; Waterson, A.G.; Lee, T.; Rossanese, O.W.; Fesik, S.W. Discovery of small molecules that bind to K-Ras and inhibit SOS-mediated activation **. Angew. Chem. 2012, 51, 6140–6143. [Google Scholar] [CrossRef] [PubMed]
- Welsch, M.E.; Kaplan, A.; Chambers, J.M.; Olive, K.P.; Ferrando, A.; Stockwell, B.R.; Sanchez-martin, M.; Badgley, M.A.; Huang, C.S.; Tran, T.H.; et al. Multivalent small-molecule Pan-RAS inhibitors. Cell 2017, 168, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Schöpel, M.; Jockers, K.F.G.; Düppe, P.M.; Autzen, J.; Potheraveedu, V.N.; Ince, S.; Yip, K.T.; Heumann, R.; Herrmann, C.; Scherkenbeck, J.; et al. Bisphenol A binds to Ras proteins and competes with guanine nucleotide exchange: Implications for GTPase-selective antagonists. J. Med. Chem. 2013, 56, 9664–9672. [Google Scholar] [CrossRef] [PubMed]
- Schöpel, M.; Herrmann, C.; Scherkenbeck, J.; Stoll, R. The Bisphenol A analogue Bisphenol S binds to K-Ras4B—Implications for “BPA-free” plastics. FEBS Lett. 2016, 590, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Martin, J.W. Prolonged exposure to Bisphenol A from single dermal contact events. Environ. Sci. Technol. 2017, 51, 9940–9949. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Spiegel, J.; Grossmann, T.N.; Waldmann, H. Direct modulation of small GTPase activity and function. Angew. Chem. Int. Ed. 2015, 54, 13516–13537. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Phan, J.; Friberg, A.R.; Camper, D.V.; Olejniczak, E.T.; Fesik, S.W. A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand. J. Biomol. NMR 2014, 60, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zou, J.; Tian, F.; Shang, Z. Fluorine bonding--how does it work in protein-ligand interactions? J. Chem. Inf. Model. 2009, 49, 2344–2355. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Boriack-Sjodin, P.A.; Margarit, S.M.; Bar-Sagi, D.; Kuriyan, J. The structural basis of the activation of Ras by SOS. Nature 1998, 394, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Nassar, N.; Horn, G.; Herrmann, C.; Block, C.; Janknecht, R.; Wittinghofer, A. Ras/Rap effector specificity determined by charge reversal. Nat. Struct. Biol. 1996, 3, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, C.; Horn, G.; Spaargaren, M.; Wittinghofer, A.; Herrmann, C.; Spaargaren, M.; Wittinghofer, A. Differential interaction of the Ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the Putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 1996, 271, 6794–6800. [Google Scholar] [CrossRef] [PubMed]
- Wittinghofer, A.; Vetter, I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011, 80, 943–971. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, C.; Martin, G.A.; Wittinghofer, A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 1995, 270, 2901–2905. [Google Scholar] [CrossRef] [PubMed]
- Kubinyi, H. Hydrogen bonding: The last mystery in drug design? Pharmacokinet. Optim. Drug Res. 2001, 513–524. [Google Scholar] [CrossRef]
- Smith, M.J.; Neel, B.G.; Ikura, M. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. USA 2013, 110, 4574–4579. [Google Scholar] [CrossRef] [PubMed]
- Rosnizeck, I.C.; Graf, T.; Spoerner, M.; Tränkle, J.; Filchtinski, D.; Herrmann, C.; Gremer, L.; Vetter, I.R.; Wittinghofer, A.; König, B.; et al. Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew. Chem. Int. Ed. 2010, 49, 3830–3833. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Yamasaki, K.; Iwahara, J.; Terada, T.; Kamiya, A.; Shirouzu, M.; Muto, Y.; Kawai, G.; Yokoyama, S.; Laue, E.D.; et al. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 1997, 36, 9109–9119. [Google Scholar] [CrossRef] [PubMed]
- Ehrkamp, A.; Herrmann, C.; Stoll, R.; Heumann, R. Ras and rheb signaling in survival and cell death. Cancers 2013, 5, 639–661. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.P.; Peters, H.; Blümer, J.; Blankenfeldt, W.; Goody, R.S.; Itzen, A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 2010, 329, 946–949. [Google Scholar] [CrossRef] [PubMed]
- Luitz, M.P.; Bomblies, R.; Ramcke, E.; Itzen, A.; Zacharias, M. Adenylylation of Tyr77 stabilizes Rab1b GTPase in an active state: A molecular dynamics simulation analysis. Sci. Rep. 2016, 6, 19896. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.; Saito, S. Relation between the conformational heterogeneity and reaction cycle of Ras: Molecular simulation of Ras. Biophys. J. 2010, 99, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Tucker, J.; Sczakiel, G.; Feuerstein, J.; John, J.; Goody, R.S.; Wittinghofer, A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986, 5, 1351–1358. [Google Scholar] [PubMed]
- Löffek, S.; Hurskainen, T.; Jackow, J.; Sigloch, F.C.; Schilling, O.; Tasanen, K.; Bruckner-Tuderman, L.; Franzke, C.W. Transmembrane collagen XVII modulates integrin dependent keratinocyte migration via PI3K/Rac1 signaling. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]

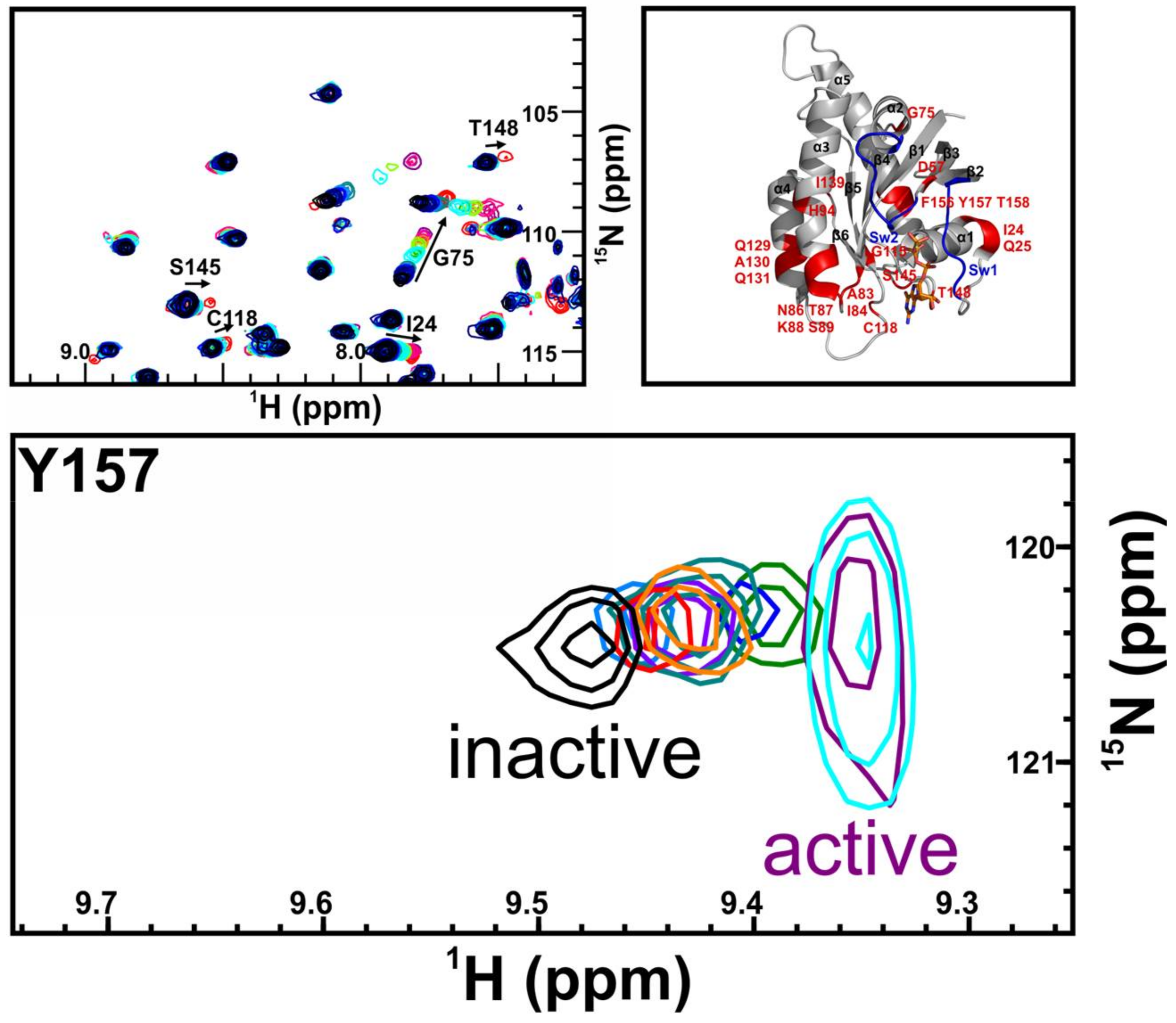
| Bisphenols tested | ||
|---|---|---|
AFX | BPF | BPE |
| KD = ND | KD = 14 ± 2 mM | KD = 6.5 ± 0.7 mM |
BPA | BPB | BPAF |
| KD = 0.6 ± 0.2 mM | KD = 3.6 ± 0.7 mM | KD = 0.4 ± 0.1 mM |
BPS | BPNH2 | BPM/BPP BPZ BPAP BPBP BPFL BPC BPP |
| KD = 5.8 ± 0.7 mM | KD = 0.4 ± 0.1 mM | Low solubility |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schöpel, M.; Shkura, O.; Seidel, J.; Kock, K.; Zhong, X.; Löffek, S.; Helfrich, I.; Bachmann, H.S.; Scherkenbeck, J.; Herrmann, C.; et al. Allosteric Activation of GDP-Bound Ras Isoforms by Bisphenol Derivative Plasticisers. Int. J. Mol. Sci. 2018, 19, 1133. https://doi.org/10.3390/ijms19041133
Schöpel M, Shkura O, Seidel J, Kock K, Zhong X, Löffek S, Helfrich I, Bachmann HS, Scherkenbeck J, Herrmann C, et al. Allosteric Activation of GDP-Bound Ras Isoforms by Bisphenol Derivative Plasticisers. International Journal of Molecular Sciences. 2018; 19(4):1133. https://doi.org/10.3390/ijms19041133
Chicago/Turabian StyleSchöpel, Miriam, Oleksandr Shkura, Jana Seidel, Klaus Kock, Xueyin Zhong, Stefanie Löffek, Iris Helfrich, Hagen S. Bachmann, Jürgen Scherkenbeck, Christian Herrmann, and et al. 2018. "Allosteric Activation of GDP-Bound Ras Isoforms by Bisphenol Derivative Plasticisers" International Journal of Molecular Sciences 19, no. 4: 1133. https://doi.org/10.3390/ijms19041133