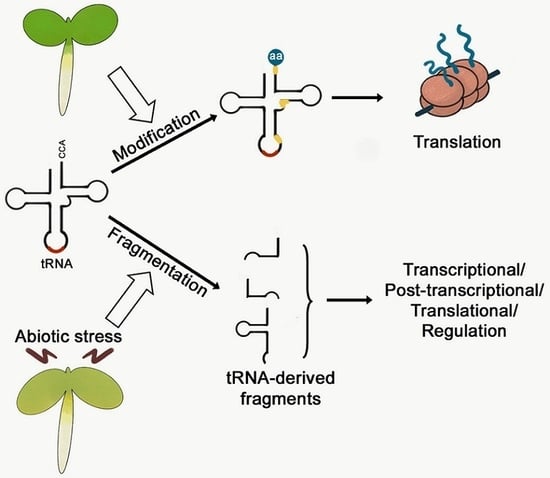
Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction
Abstract
1. Introduction
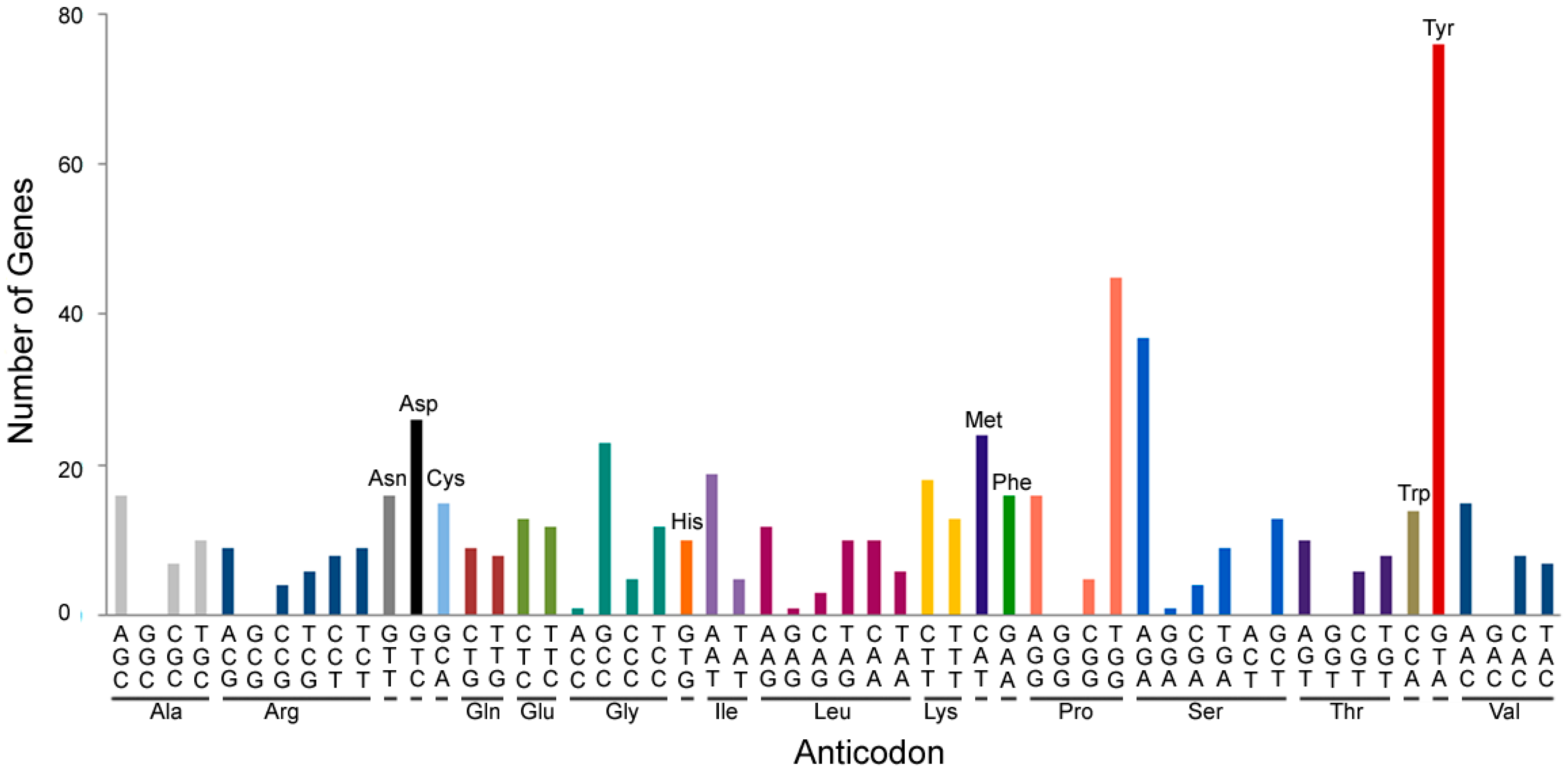
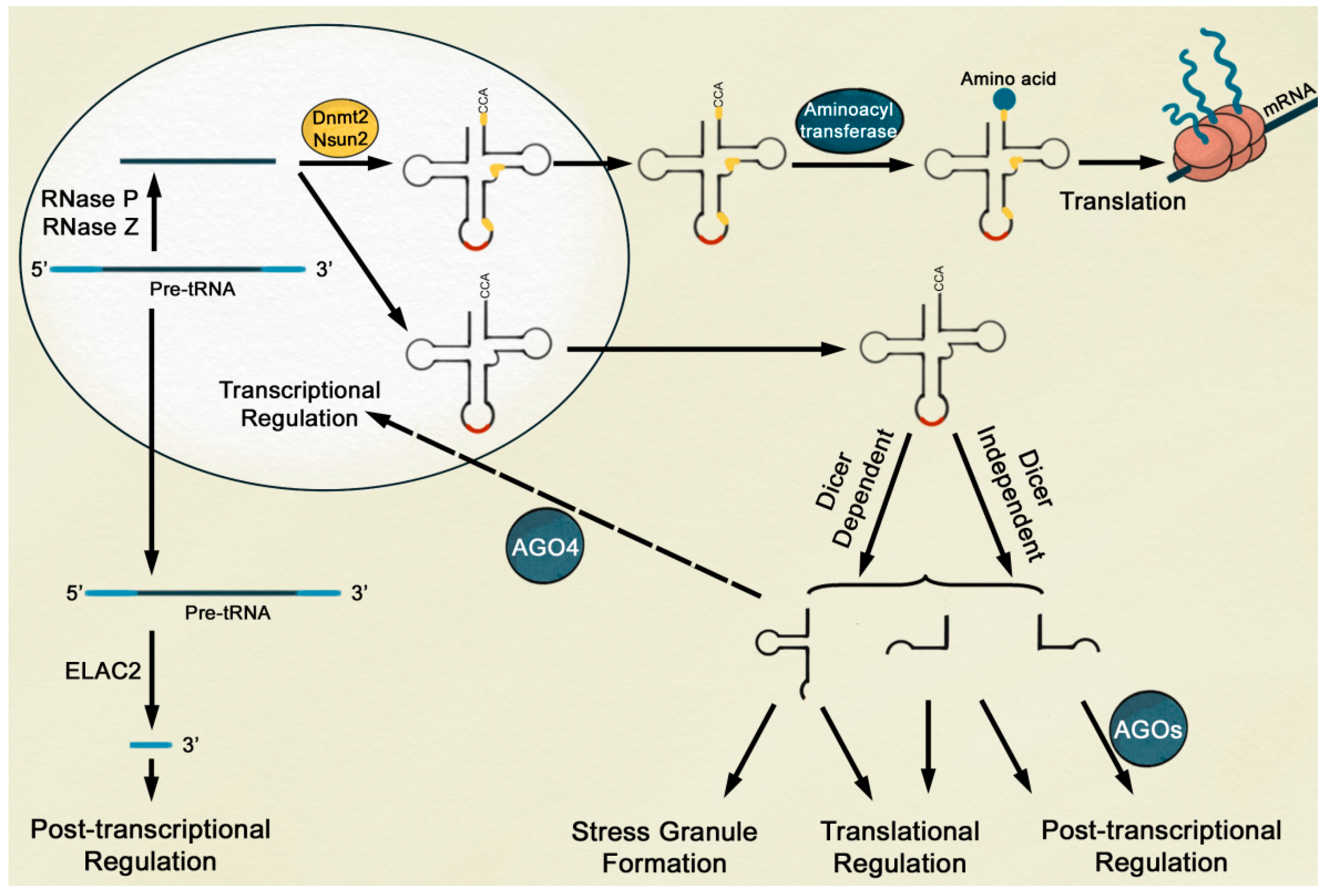
2. tRNA-Derived Fragments Are Small ncRNAs with a Specific Function
3. Biogenesis of tRFs Possibly Occurs through Dicer-Dependent and -Independent Pathways
4. tRFs Are Involved in Post-Transcriptional and Translational Gene Silencing
5. Function of tRFs during Abiotic Stress Signal Transduction in Plants
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Bohmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Nguyen, A.H.; Nakaminami, K.; Seki, M. Arabidopsis non-coding RNA regulation in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 22642–22654. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Jager, G.; Zheng, B. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 201. [Google Scholar] [CrossRef] [PubMed]
- Megel, C.; Morelle, G.; Lalande, S.; Duchene, A.M.; Small, I.; Marechal-Drouard, L. Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 2015, 16, 1873–1893. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. Stressing out over tRNA cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.S.; Vicentini, R.; Duarte, G.T.; Pinoti, V.F.; Vincentz, M.; Nogueira, F.T. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol. Biol. 2017, 93, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Liu, Q.; Zhang, Y.C.; Qu, L.H.; Chen, Y.Q.; Gautheret, D. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol. 2011, 8, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013, 14, 298. [Google Scholar] [CrossRef] [PubMed]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013, 194, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Kawamura, T.; Loher, P.; Morichika, K.; Rigoutsos, I.; Kirino, Y. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017, 45, 9108–9120. [Google Scholar] [CrossRef] [PubMed]
- Olvedy, M.; Scaravilli, M.; Hoogstrate, Y.; Visakorpi, T.; Jenster, G.; Martens-Uzunova, E.S. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 2016, 7, 24766–24777. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Sun, Q.; Yao, Y. Characterization of Small RNAs Derived from tRNAs, rRNAs and snoRNAs and Their Response to Heat Stress in Wheat Seedlings. PLoS ONE 2016, 11, e0150933. [Google Scholar] [CrossRef] [PubMed]
- Loss-Morais, G.; Waterhouse, P.M.; Margis, R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct 2013, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Asman, A.K.; Vetukuri, R.R.; Jahan, S.N.; Fogelqvist, J.; Corcoran, P.; Avrova, A.O.; Whisson, S.C.; Dixelius, C. Fragmentation of tRNA in Phytophthora infestans asexual life cycle stages and during host plant infection. BMC Microbiol. 2014, 14, 308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, T.; Xu, K.; Zhang, W.; Wang, X.; Quan, J.; Jin, W.; Zhang, M.; Fan, G.; Wang, M.B.; et al. The tRNA-Derived Small RNAs Regulate Gene Expression through Triggering Sequence-Specific Degradation of Target Transcripts in the Oomycete Pathogen Phytophthora sojae. Front. Plant Sci. 2016, 7, 1938. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Choudury, S.G.; Slotkin, R.K. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017, 45, 5142–5152. [Google Scholar] [CrossRef] [PubMed]
- Cognat, V.; Morelle, G.; Megel, C.; Lalande, S.; Molinier, J.; Vincent, T.; Small, I.; Duchene, A.M.; Marechal-Drouard, L. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 2016, 45, 3460–3472. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.K.; Rybak, S.M.; Davey, R.T., Jr.; Youle, R.J.; Ackerman, E.J. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J. Biol. Chem. 1992, 267, 21982–21986. [Google Scholar] [PubMed]
- Thiagarajan, D.; Dev, R.R.; Khosla, S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics 2011, 6, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.L.; David, R.; Searle, I.R. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015, 15, 199. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, I.; Hansen, T.B. Biogenesis and Function of Ago-Associated RNAs. Trends Genet. 2017, 33, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Karaiskos, S.; Grigoriev, A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Research 2016, 24. [Google Scholar] [CrossRef] [PubMed]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.; Ham, J.; Jang, E.S.; Chi, S.W. MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions. Mol. Cells 2016, 39, 375–381. [Google Scholar] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Bounova, G.; Purdom, E.; Speed, T.P.; Collins, K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell 2012, 48, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Sachidanandam, R.; Collins, K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010, 24, 2742–2747. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Lee, S.R.; Hogstad, B.; Malone, C.D.; Tonkin, L.A.; Sachidanandam, R.; Hannon, G.J.; Collins, K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009, 23, 2016–2032. [Google Scholar] [CrossRef] [PubMed]
- Hock, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Pang, Y.L.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Zywicki, M.; Kunzi, A.; Polacek, N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef] [PubMed]
- Hillwig, M.S.; Lebrasseur, N.D.; Green, P.J.; Macintosh, G.C. Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Mol. Genet. Genom. 2008, 280, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pang, C.; Li, X.; Hu, Z.; Lv, Z.; Zheng, B.; Chen, P. Identification of tRNA nucleoside modification genes critical for stress response and development in rice and Arabidopsis. BMC Plant Biol. 2017, 17, 261. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wu, K.; Dhaubhadel, S.; An, L.; Tian, L. Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochem. Biophys. Res. Commun. 2010, 396, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Sridha, S.; Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006, 46, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, Y.Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lee, I.; Ren, J.; Ajay, S.S.; Lee, Y.S.; Bao, X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013, 21, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Shi, J.; Zhang, H.; Cao, Z.; Gao, X.; Ren, W.; Ning, Y.; Ning, L.; Cao, Y.; et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 2014, 6, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, L.; Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009, 150, 378–387. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.J.; Kim, T.-H. Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction. Int. J. Mol. Sci. 2018, 19, 518. https://doi.org/10.3390/ijms19020518
Park EJ, Kim T-H. Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction. International Journal of Molecular Sciences. 2018; 19(2):518. https://doi.org/10.3390/ijms19020518
Chicago/Turabian StylePark, Eun Joo, and Tae-Houn Kim. 2018. "Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction" International Journal of Molecular Sciences 19, no. 2: 518. https://doi.org/10.3390/ijms19020518
APA StylePark, E. J., & Kim, T.-H. (2018). Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction. International Journal of Molecular Sciences, 19(2), 518. https://doi.org/10.3390/ijms19020518