Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance
Abstract
:1. Introduction
2. Results
2.1. Molecular Characterization of Posox9a and Posox9b
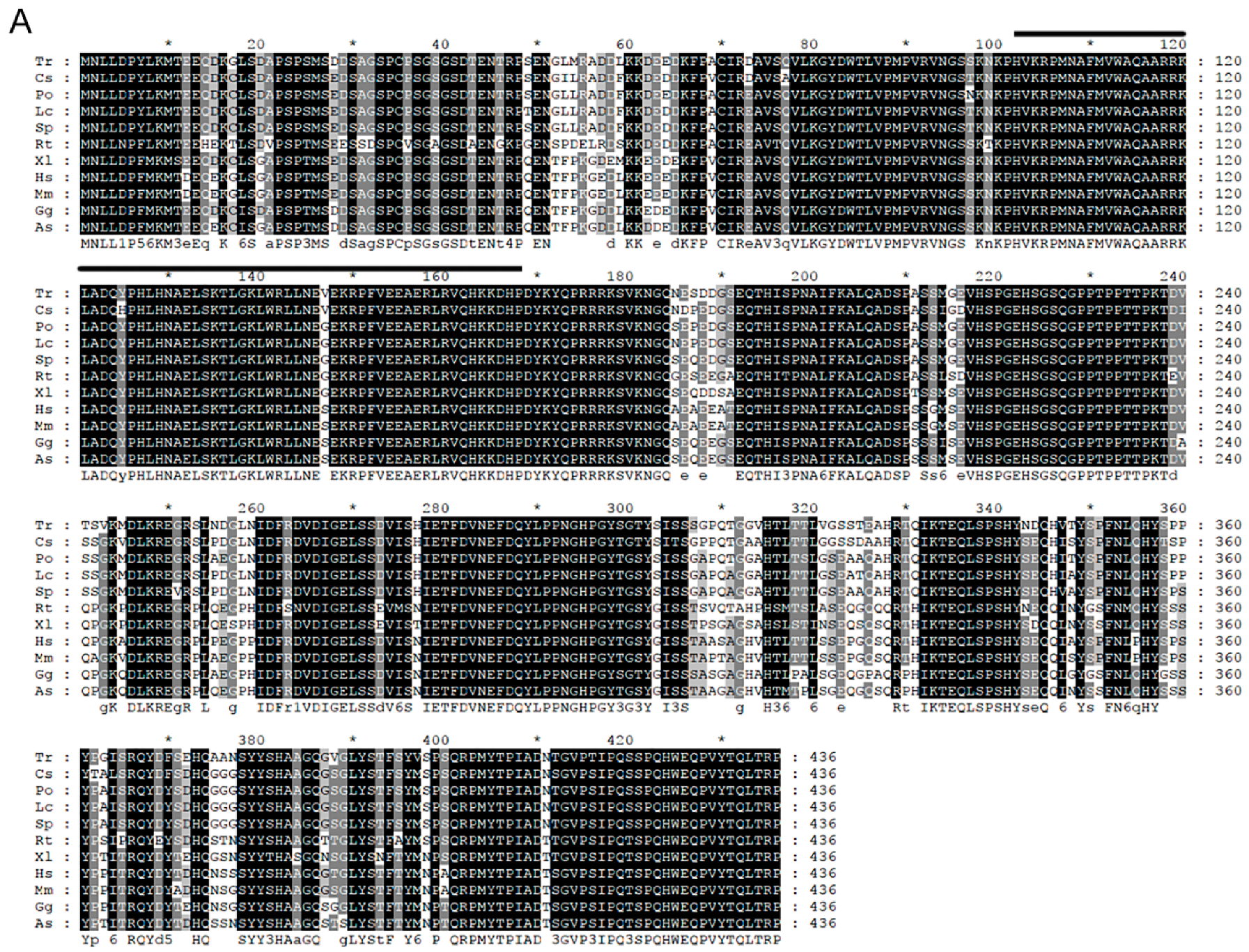
2.1.1. Cloning and Sequence Analysis of Posox9a and Posox9b
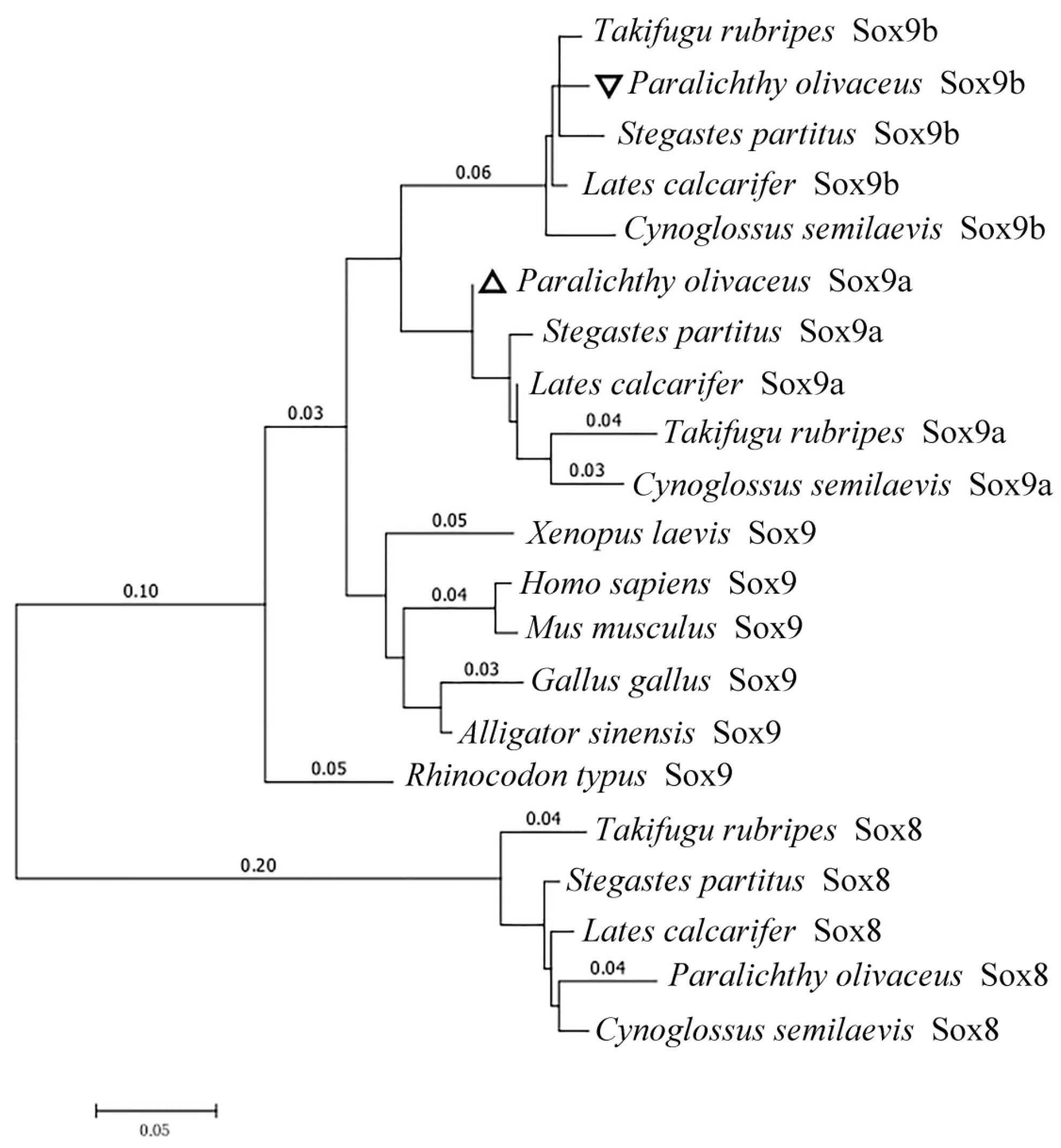
2.1.2. Homology and Phylogenetic Analysis
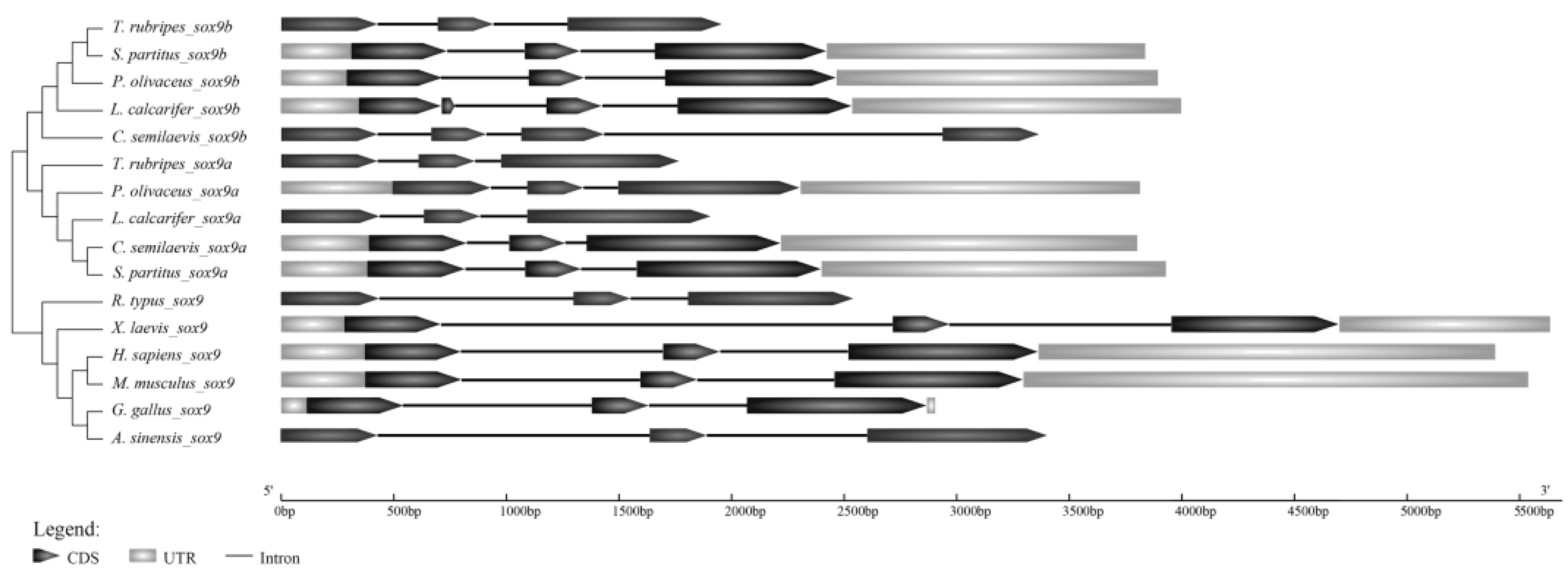
2.1.3. Genomic Structure of Posox9 Genes
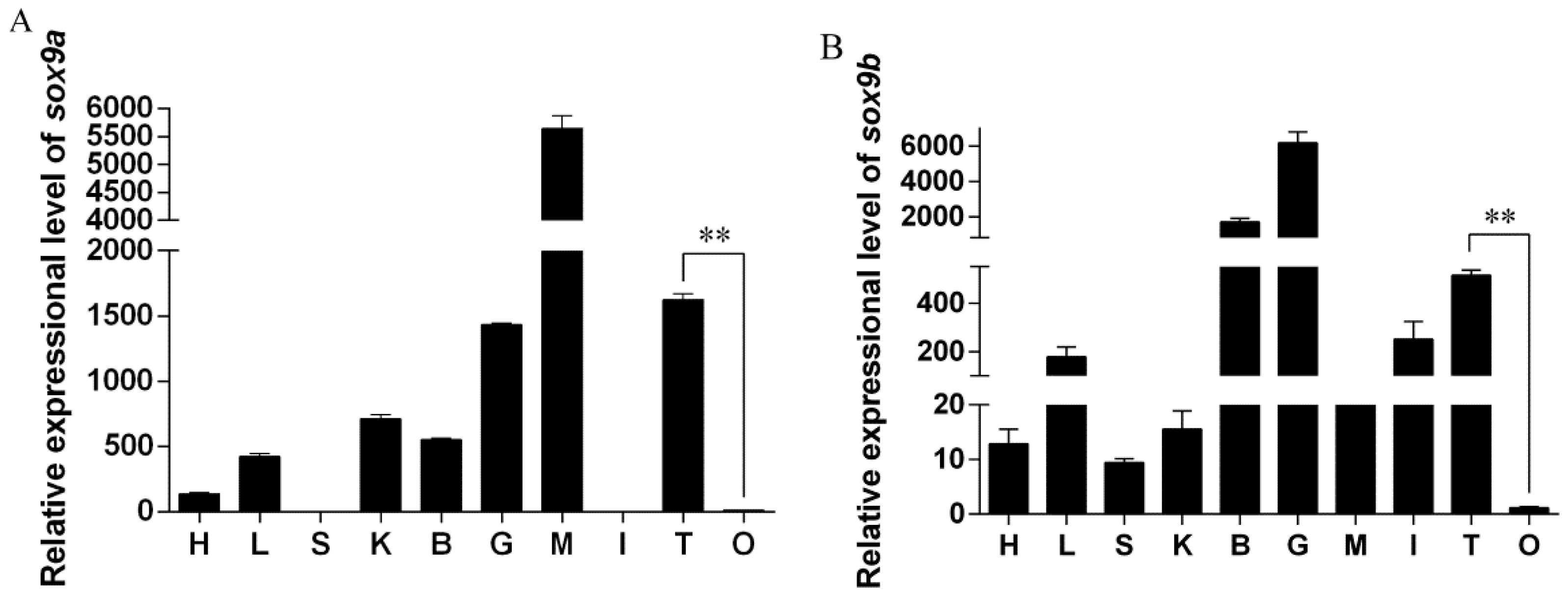
2.2. Relative Expression of Posox9a and Posox9b in Different Tissues
2.3. Sexually Dimorphic Expression of Posox9a and Posox9b Mrnas in Gonads
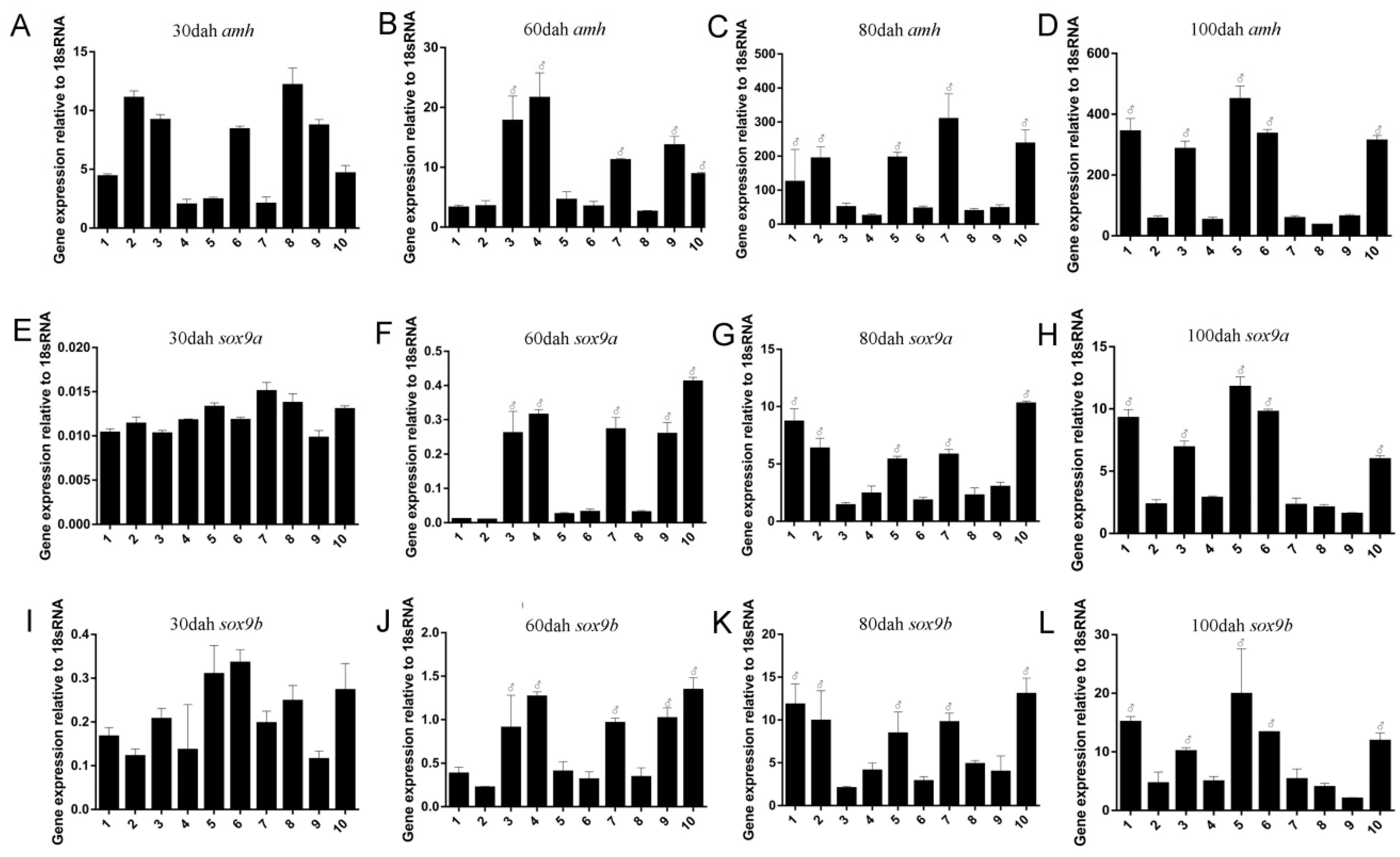
2.4. Expression Profile of Posox9a and Posox9b after 17α-methyltestosterone Injection
3. Discussion
3.1. Posox9a and Posox9b Are Highly Conversed in Vertebrates
3.2. Both Posox9a and Posox9b Are More Abundant in Testis Than in Ovary
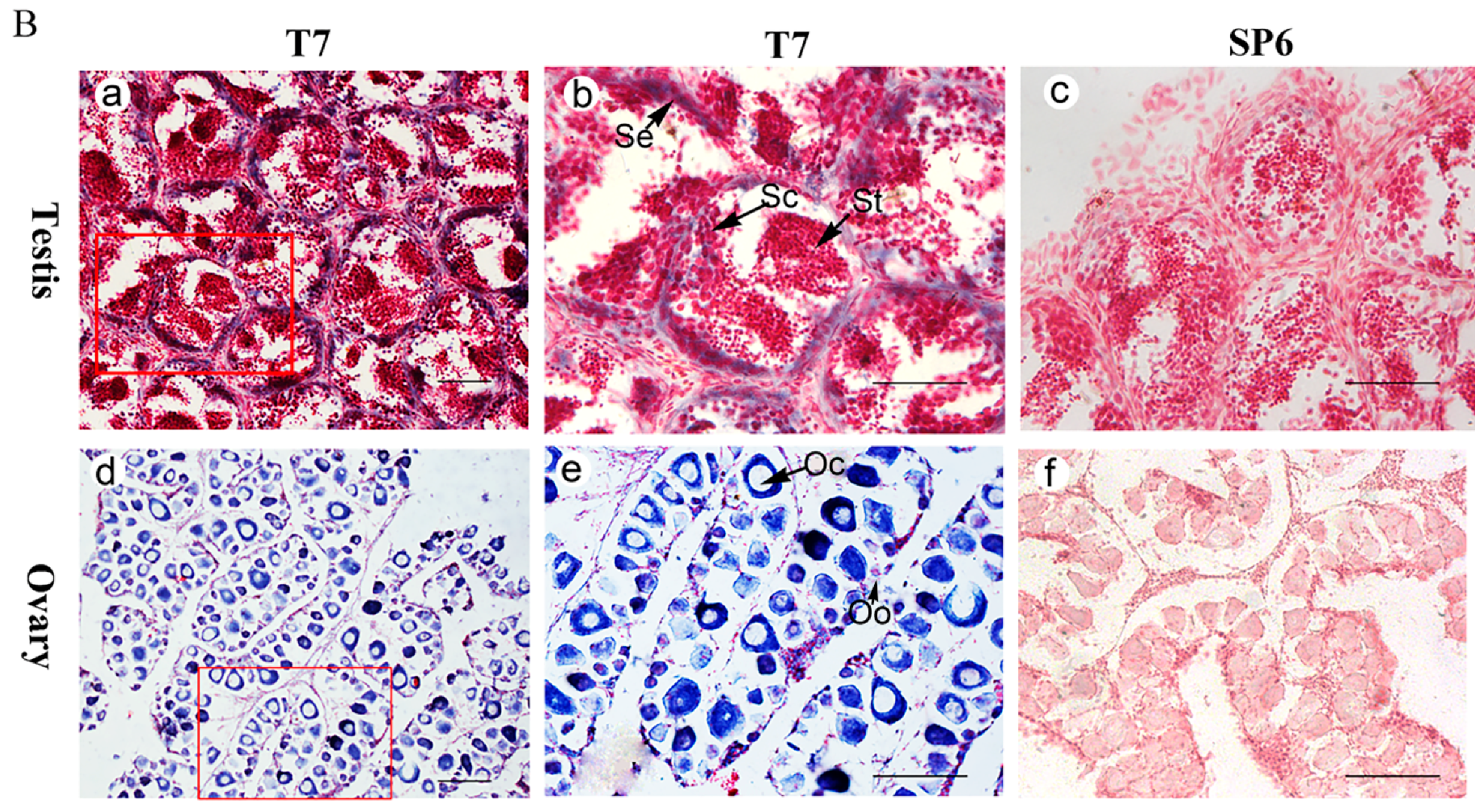
3.3. Posox9a and Posox9b Transcripts Are Detected in Sertoli Cells, Spermatocytes and Oocytes
3.4. Sexually Dimorphic Expression of Posox9a and Posox9b Mrnas in Gonads during Sex Differentiation
3.5. 17α-methyltestosterone Increased the Expression of Posox9b but Not Posox9a
4. Materials and Methods
4.1. Ethics Statement
4.2. Fish and Sampling
4.3. RNA Extraction and cDNA Synthesis
4.4. Molecular Cloning of Posox9a and Posox9b
4.5. Sequence Analysis
4.6. Quantitative Real-Time PCR (qRT-PCR)
4.7. ISH
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gubbay, J.; Collignon, J.; Koopman, P.; Capel, B.; Economou, A.; Münsterberg, A.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R.; Gubbay, J.; et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a new family of embryonically expressed genes. Nature 1990, 346, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovellbadge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef] [PubMed]
- She, Z.Y.; Yang, W.X. Sox family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015, 94, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Yi, W. The relationship between Sox9 gene and sex determination. Chem. Life Chin. 2000, 20, 70–71. [Google Scholar]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Healy, C.; Uwanogho, D.; Sharpe, P.T. Regulation and role of Sox9 in cartilage formation. Dev. Dyn. 1999, 215, 69–78. [Google Scholar] [CrossRef]
- Meyer, J. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the sry-related gene Sox9. Cell 1994, 79, 1111–1120. [Google Scholar]
- She, Z.Y.; Yang, W.X.; Yang, W.X. Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 2016, 63, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kent, J.; Wheatley, S.C.; Andrews, J.E.; Sinclair, A.H.; Koopman, P. A male-specific role for Sox9 in vertebrate sex determination. Development 1996, 122, 2813–2822. [Google Scholar] [PubMed]
- Morais, d.S.S.; Hacker, A.; Harley, V.; Goodfellow, P.; Swain, A.; Lovellbadge, R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat. Genet. 1996, 14, 62–68. [Google Scholar]
- Moreno-Mendoza, N.; Harley, V.R.; Merchant-Larios, H. Differential expression of Sox9 in gonads of the sea turtle lepidochelys olivacea at male-or female-promoting temperatures. J. Exp. Zool. 1999, 284, 705–710. [Google Scholar] [CrossRef]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two Sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cui, J.; Yang, G.; Gong, Q.; Gu, Q. Expression detection of dmrts and two Sox 9 genes in takifugu rubripes (tetraodontidae, vertebrata). J. Ocean Univ. China 2007, 6, 182–186. [Google Scholar] [CrossRef]
- Smith, E.K.; Guzmán, J.M.; Luckenbach, J.A. Molecular cloning, characterization, and sexually dimorphic expression of five major sex differentiation-related genes in a scorpaeniform fish, sablefish (Anoplopoma fimbria). Comp. Biochem. Physiol. Part B 2013, 165, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Spotila, L.D.; Spotila, J.R.; Hall, S.E. Sequence and expression analysis of wt1 and Sox9 in the red-eared slider turtle, Trachemys scripta. J. Exp. Zool. 1998, 281, 417–427. [Google Scholar] [CrossRef]
- Stolt, C.C.; Lommes, P.; Sock, E.; Chaboissier, M.C.; Schedl, A.; Wegner, M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003, 17, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Spokony, R.F.; Aoki, Y.; Saint-Germain, N.; Magner-Fink, E.; Saint-Jeannet, J.-P. The transcription factor sox9 is required for cranial neural crest development in xenopus. Development 2002, 129, 421–432. [Google Scholar] [PubMed]
- Yamaguchi, T.; Yoshinaga, N.; Yazawa, T.; Gen, K.; Kitano, T. Cortisol is involved in temperature-dependent sex determination in the japanese flounder. Endocrinology 2010, 151, 3900–3908. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Shiraishi, E.; Yamamoto, T.; Iguchi, T.; Abe, S.-I.; Kitano, T. Sexually dimorphic expression of a teleost homologue of müllerian inhibiting substance during gonadal sex differentiation in japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2004, 322, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Du, X.; Zhao, J.; Liu, X.; Li, X.; Zhang, Q.; Wang, X. Sexually dimorphic expression in developing and adult gonads shows an important role of gonadal soma-derived factor during sex differentiation in olive flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part B 2017, 210, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, F.; Georg, I.; Scherthan, H.; Lécureuil, C.; Guillou, F.; Wegner, M.; Scherer, G. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev. Biol. 2009, 327, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Chen, S.; Ji, X.; Shao, C. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin. 2011, 30, 68–77. [Google Scholar] [CrossRef]
- Raghuveer, K.; Senthilkumaran, B. Isolation of Sox9 duplicates in catfish: Localization, differential expression pattern during gonadal development and recrudescence, and hcg-induced up-regulation of Sox9 in testicular slices. Reproduction 2010, 140, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Gorczynska, E.; Handelsman, D. The role of calcium in follicle-stimulating hormone signal transduction in Sertoli cells. J. Biol. Chem. 1991, 266, 23739–23744. [Google Scholar] [PubMed]
- French, F.; Ritzen, E. Androgen-binding protein in efferent duct fluid of rat testis. J. Reprod. Fertil. 1973, 32, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H. Gonadal sex differentiation in flounder, Paralichthys olivaceus. Bull. Natl. Res. Inst. Aquac. 1987, 11, 7–19. [Google Scholar]
- Yamamoto, E. Studies on sex-manipulation and production of cloned populations in hirame, Paralichthys olivaceus (temminck et schlegel). Aquaculture 1999, 173, 235–246. [Google Scholar] [CrossRef]
- Kitano, T.; Takamune, K.; Nagahama, Y.; Abe, S.-I. Aromatase inhibitor and 17a-methyltestosterone cause sex-reversal from genetical females to phenotypic males and suppression of p450 aromatase gene expression in japanese flounder (Paralichthys olivaceus). Mol. Reprod. Dev. 2000, 56, 1–5. [Google Scholar] [CrossRef]
- Josso, N.; Cate, R.; Picard, J.; Vigier, B.; di Clemente, N.; Wilson, C.; Imbeaud, S.; Pepinsky, R.; Guerrier, D.; Boussin, L. Anti-müllerian hormone: The jost factor. Recent Prog. Horm. Res. 1993, 48, 1–59. [Google Scholar] [PubMed]
- Hoar, W.S.; Randall, D.J.; Donaldson, E.M. Fish Physiology; Academic Press: Cambridge, MA, USA, 1983. [Google Scholar]
- Fisher, A.; Horsfield, J.A.; Rutherford, K.; Black, M.A.; Gemmell, N.J.; Lee, S.L.J. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad development. BMC Genom. 2017, 18, 557. [Google Scholar]
- Wang, W.; Wang, J.; You, F.; Ma, L.; Yang, X.; Gao, J.; He, Y.; Qi, J.; Yu, H.; Wang, Z. Detection of alternative splice and gene duplication by rna sequencing in japanese flounder, Paralichthys olivaceus. G3 2014, 4, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, J.; Jiang, J.; Fan, L.; Wang, W.; Liu, J.; Zhang, Q.; Wang, X. Identification and characterization of a nanog homolog in japanese flounder (Paralichthys olivaceus). Gene 2013, 531, 411–421. [Google Scholar] [CrossRef] [PubMed]



| Period | 30 Dah | 40 Dah | 50 Dah | 60 Dah | 70 Dah | 80 Dah | 90 Dah | 100 Dah |
|---|---|---|---|---|---|---|---|---|
| TL (mm) | 11.4 | 16.6 | 24.1 | 32.7 | 43.6 | 50.4 | 54.9 | 61.4 |
| Primer Name | Sequence (5′–3′) | Usage |
|---|---|---|
| Posox9a-Fw | ATGAATCTCCTCGACCCTTACC | ORF amplification |
| Posox9a-Rv | TCAGGGTCTGGTGAGCTGG | ORF amplification |
| Posox9a-qRT-Fw | AACGAAGGCGAGAAGCGG | qRT-PCR |
| Posox9a-qRT-Rv | ATGGCATTAGGAGAAATGTGCG | qRT-PCR |
| Posox9a-ISH-Fw | AATCCTCTGTGAGGACTTATTG | ISH probe |
| Posox9a-ISH-Rv | CAGATTCCGTCTCTCTTTCTC | ISH probe |
| Posox9b-Fw | ATCTGTGATACGCTGTTCTTT | ORF amplification |
| Posox9b-Rv | CTCTTCACGGCCTGGAC | ORF amplification |
| Posox9b-qRT-Fw | GCGAGCAAACGCACATA | qRT-PCR |
| Posox9b-qRT-Rv | CCAAAGTCGATGTTGAGCTG | qRT-PCR |
| Posox9b-ISH-Fw | CTTTGAATTGGCTCACAACAG | ISH probe |
| Posox9b-ISH-Rv | ACTGATGACACGGTCATATC | ISH probe |
| amh-qRT-FW | AGCACTGACAGTTTCTCATCC | qRT-PCR |
| amh-qRT-RV | GTAAGACTGATCCCGATGAACTG | qRT-PCR |
| 18s-qRT-FW | GGTAACGGGGAATCAGGGT | qRT-PCR |
| 18s-qRT-RV | TGCCTTCCTTGGATGTGGT | qRT-PCR |
| β-actin-qRT-Fw | GAGATGAAGCCCAGAGCAAGAG | qRT-PCR |
| β-actin-qRT-Rv | CAGCTGTGGTGGTGAAGGAGTAG | qRT-PCR |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yu, H.; Wang, Y.; Liu, X.; Liu, Y.; Qu, J.; Wang, X. Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance. Int. J. Mol. Sci. 2018, 19, 512. https://doi.org/10.3390/ijms19020512
Li X, Yu H, Wang Y, Liu X, Liu Y, Qu J, Wang X. Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance. International Journal of Molecular Sciences. 2018; 19(2):512. https://doi.org/10.3390/ijms19020512
Chicago/Turabian StyleLi, Xiaojing, Haiyang Yu, Yujue Wang, Xiaobing Liu, Yuezhong Liu, Jiangbo Qu, and Xubo Wang. 2018. "Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance" International Journal of Molecular Sciences 19, no. 2: 512. https://doi.org/10.3390/ijms19020512






