Importance of a Laccase Gene (Lcc1) in the Development of Ganoderma tsugae
Abstract
:1. Introduction
2. Results
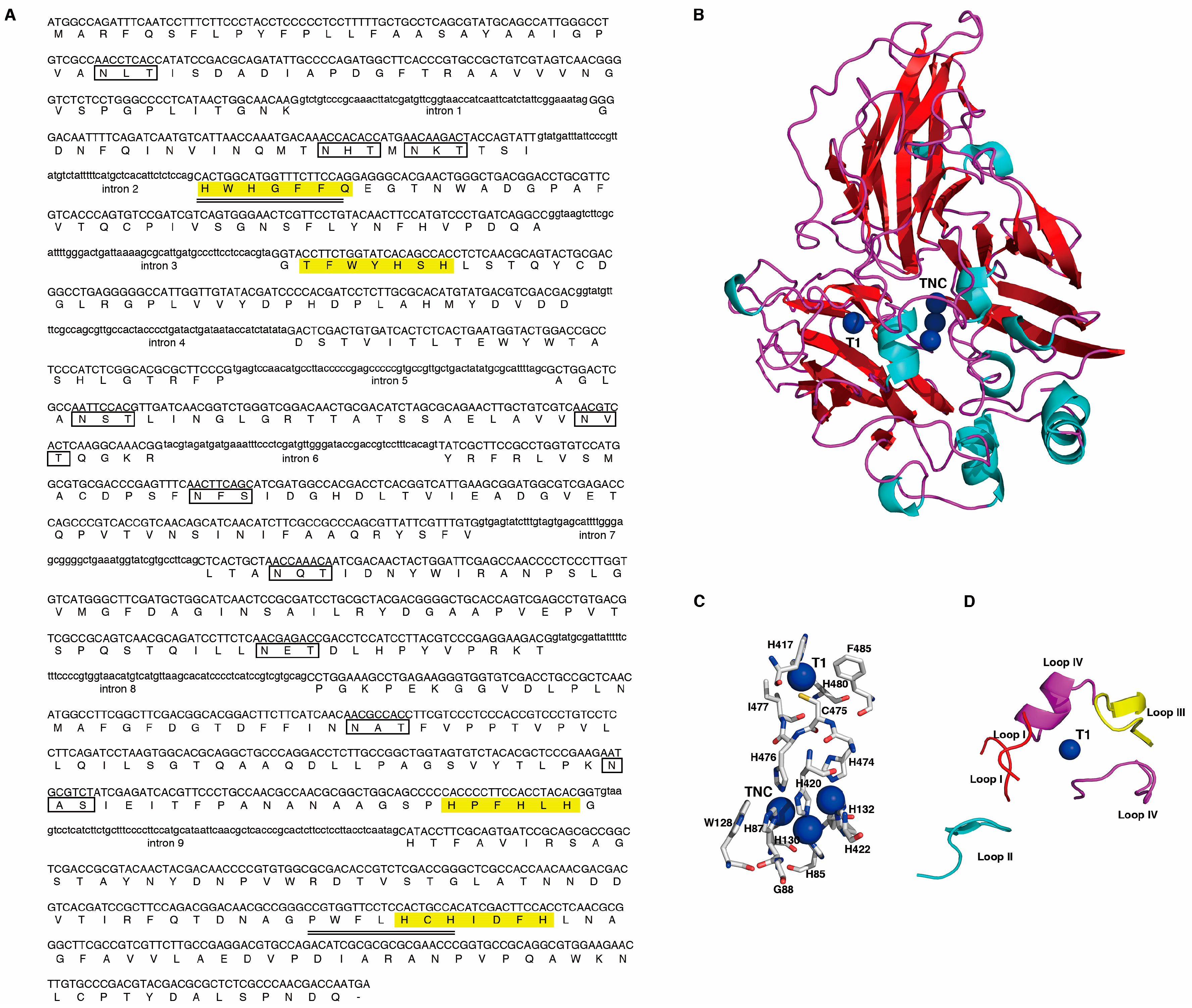
2.1. Cloning and Analysis of the Lcc1 Sequence of G. tsugae
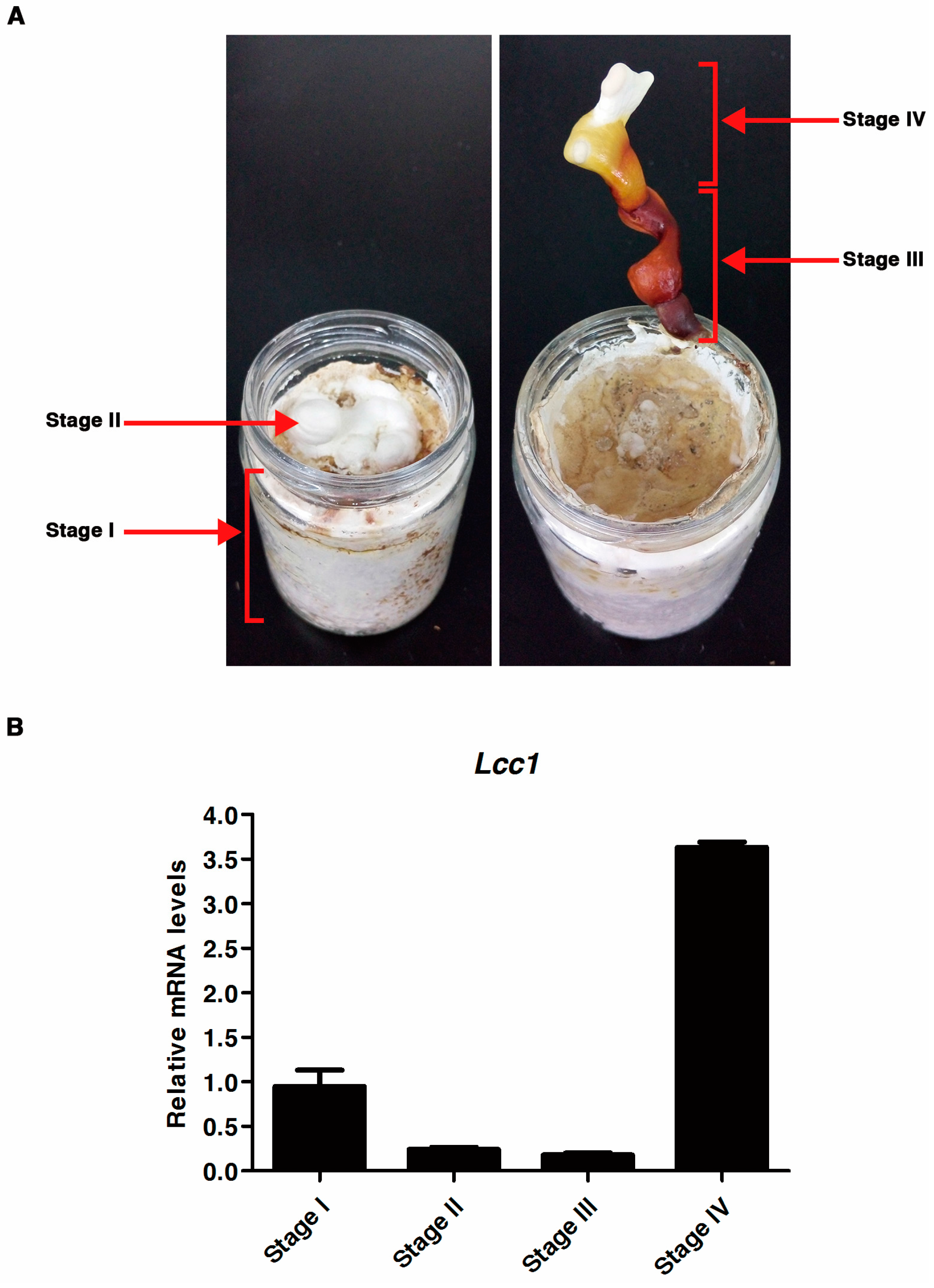
2.2. Expression Profile of Lcc1 at Different Developmental Stages of G. tsugae
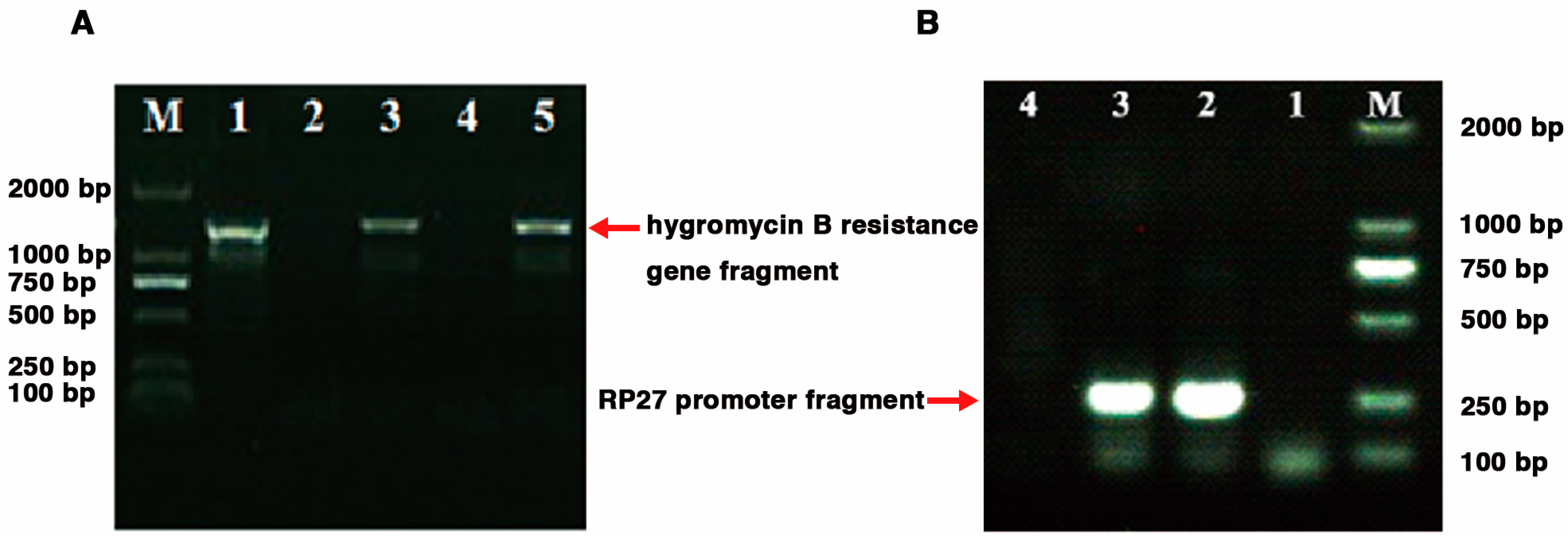
2.3. Generation of G. tsugae Lcc1 Transgenic Strains
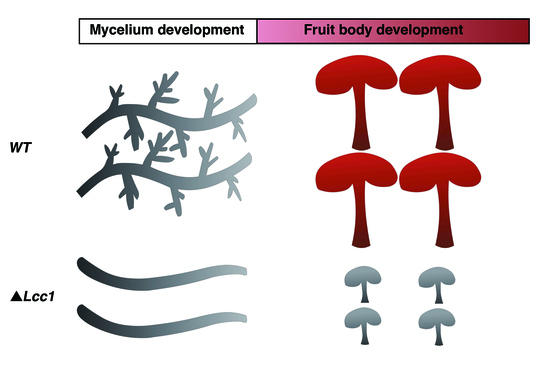
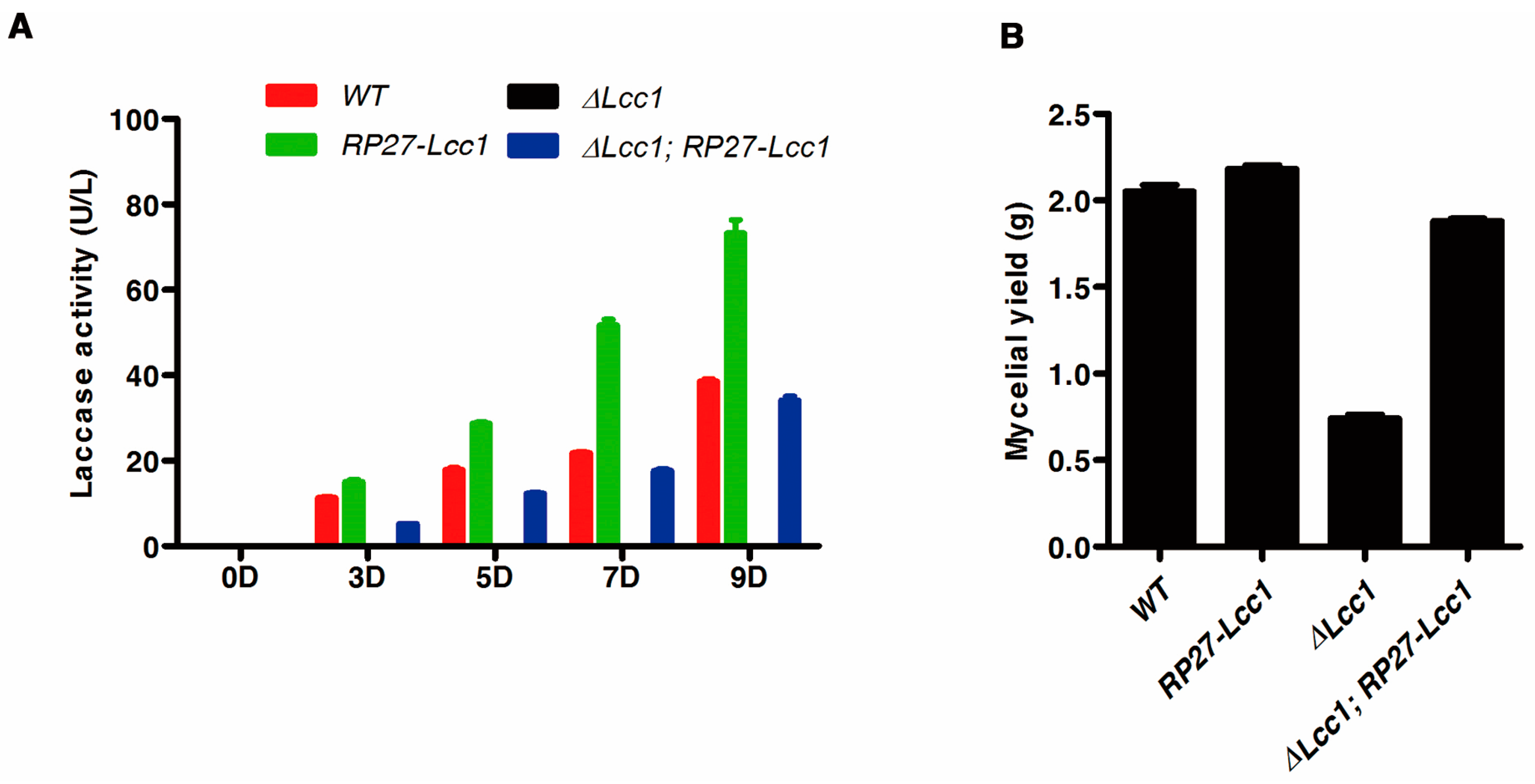
2.4. Lcc1 Is Required for Mycelium Development
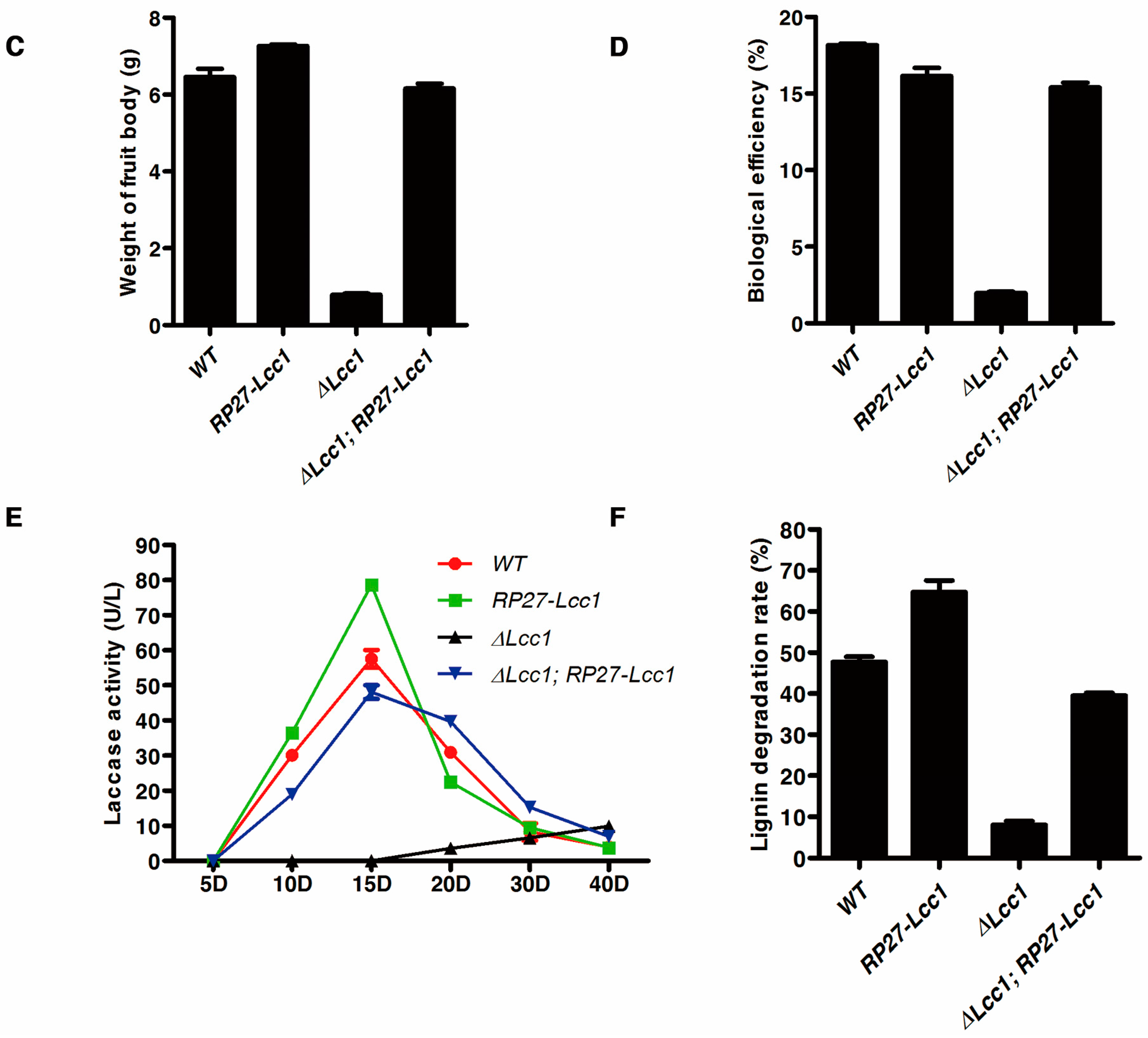
2.5. Lcc1 Is Essential for G. tsugae Development
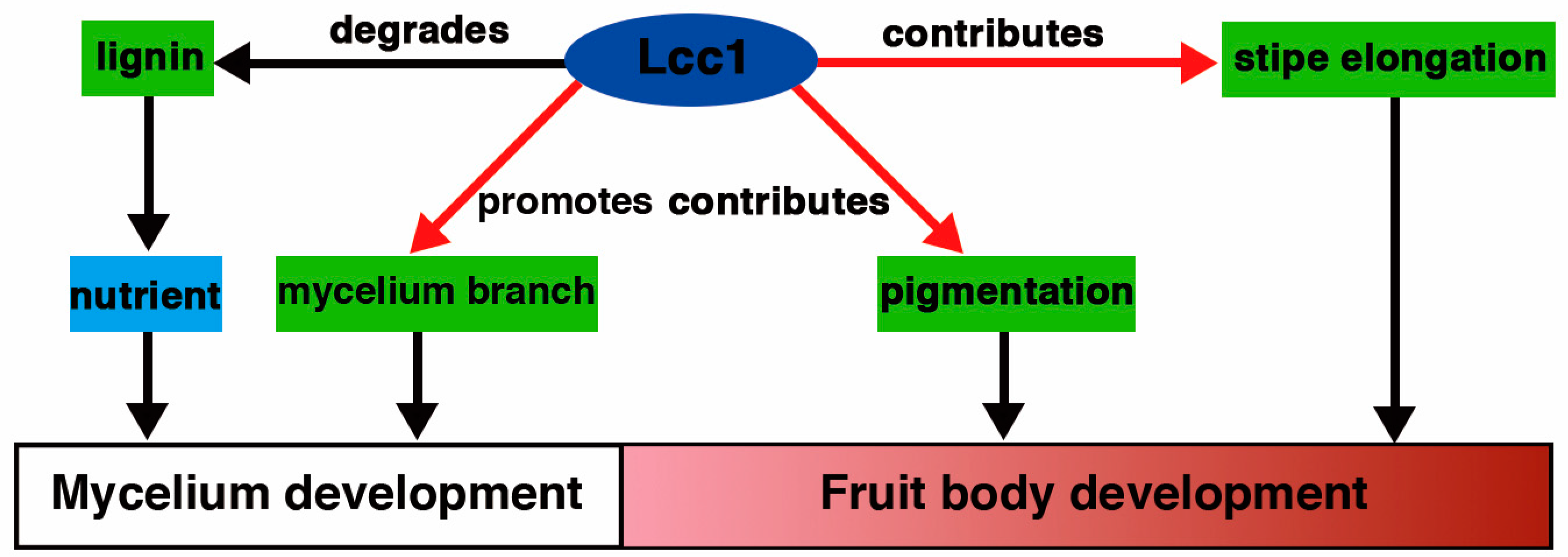
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Molecular Biology Procedures and Plasmid Constructions
4.2.1. Cloning the Lcc1 Gene from G. tsugae
4.2.2. Analysis of Lcc1 RNA Expression Levels during the G. Tsugae Life Cycle
4.2.3. Generation of Lcc1 Knockout Cassettes
4.2.4. Generation of Lcc1 Transgenic Strains by Genetic Manipulation Approaches
4.3. Enzyme Assays
4.4. Scan Electronic Microscope Analysis
4.5. Determining Lignin Degradation Rate by 72% Sulfuric Acid Method
4.6. Nucleotide Sequence Accession Number
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Patel, H.; Gupte, S.; Gupte, A. Laccases: The biocatalyst with industrial and biotechnological applications. In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B.N., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 309–342. [Google Scholar]
- Kawai, S.; Umezawa, T.; Shimada, M.; Higuchi, T. Aromatic ring cleavage of 4,6-di(tert-butyl)guaiacol, a phenolic lignin model compound, by laccase of coriolus versicolor. FEBS Lett. 1988, 236, 309–311. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.E. Laccase is essential for lignin degradation by the white-rot fungus pycnoporus cinnabarinus. FEBS Lett. 1997, 407, 89–92. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Dean, J.F.D.; Eriksson, K.E.L. Laccase-mediated formation of the phenoxazinone derivative, cinnabarinic acid. FEBS Lett. 1995, 376, 202. [Google Scholar] [CrossRef]
- Das, N.; Sengupta, S.; Mukherjee, M. Importance of laccase in vegetative growth of pleurotus florida. Appl. Environ. Microbiol. 1997, 63, 4120–4122. [Google Scholar] [PubMed]
- Wang, W.; Liu, F.; Jiang, Y.; Wu, G.; Guo, L.; Chen, R.; Chen, B.; Lu, Y.; Dai, Y.; Xie, B. The multigene family of fungal laccases and their expression in the white rot basidiomycete flammulina velutipes. Gene 2015, 563, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Aggelis, G.; Ehaliotis, C.; Nerud, F.; Stoychev, I.; Lyberatos, G.; Zervakis, G.I. Evaluation of white-rot fungi for detoxification and decolorization of effluents from the green olive debittering process. Appl. Microbiol. Biotechnol. 2002, 59, 353–360. [Google Scholar] [PubMed]
- Alcalde, M. Laccases: Biological Functions, Molecular Structure and Industrial Applications; Springer: Dordrecht, The Netherlands, 2007; pp. 461–476. [Google Scholar]
- Wang, H.X.; Ng, T.B. Purification of a novel low-molecular-mass laccase with HIV-1 reverse transcriptase inhibitory activity from the mushroom Tricholoma giganteum. Biochem. Biophys. Res. Commun. 2004, 315, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B.; Jiang, Y.; Liu, F.; Sze, S.C.; Zhang, K.Y. Purification and characterization of a laccase with inhibitory activity toward HIV-1 reverse transcriptase and tumor cells from an edible mushroom (Pleurotus cornucopiae). Protein Pept. Lett. 2010, 17, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Ng, T.B. Purification of a laccase from fruiting bodies of the mushroom pleurotus eryngii. Appl. Microbiol. Biotechnol. 2006, 69, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Ng, T.B. A laccase from the medicinal mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2006, 72, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Eggert, C.; LaFayette, P.R.; Temp, U.; Eriksson, K.E.; Dean, J.F. Molecular analysis of a laccase gene from the white rot fungus pycnoporus cinnabarinus. Appl. Environ. Microbiol. 1998, 64, 1766–1772. [Google Scholar] [PubMed]
- Wei, Y.S.; Wung, B.S.; Lin, Y.C.; Hsieh, C.W. Isolating a cytoprotective compound from ganoderma tsugae: Effects on induction of nrf-2-related genes in endothelial cells. Biosci. Biotechnol. Biochem. 2009, 73, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Baby, S.; Johnson, A.J.; Govindan, B. Secondary metabolites from ganoderma. Phytochemistry 2015, 114, 66–101. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Liu, J.Z.; Hu, K.H.; Zhu, H.X. The level of secreted laccase activity in the edible fungi and their growing cycles are closely related. Curr. Microbiol. 2011, 62, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Kenkebashvili, N.V.; Elisashvili, V.; Hadar, Y. Effect of nutrient medium composition on laccase and manganese peroxidase activity in medicinal mushrooms. Int. J. Med. Mushrooms 2009, 11, 191–198. [Google Scholar] [CrossRef]
- Sun, J.; Peng, R.H.; Xiong, A.S.; Tian, Y.S.; Zhao, W.; Xu, H.; Liu, D.T.; Chen, J.M.; Yao, Q.H. Secretory expression and characterization of a soluble laccase from the Ganoderma lucidum strain 7071-9 in Pichia pastoris. Mol. Biol. Rep. 2012, 39, 3807–3814. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.; Ma, L.; Fan, F.F.; Gong, Y.M.; Wan, X.; Jiang, M.L.; Zhang, X.Y.; Yang, Y. Decolorization of different dyes by a newly isolated white-rot fungi strain Ganoderma sp.En3 and cloning and functional analysis of its laccase gene. J. Hazard. Mater. 2011, 192, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.T.; Tai, R.; Hseu, R.S.; Huang, C.T. Overexpression and characterization of a thermostable, pH-stable and organic solvent-tolerant Ganoderma fornicatum laccase in Pichia pastoris. Process Biochem. 2011, 46, 1469–1474. [Google Scholar] [CrossRef]
- Fairhead, C.; Llorente, B.; Denis, F.; Soler, M.; Dujon, B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 1996, 12, 1439–1457. [Google Scholar] [CrossRef]
- Jeong, J.S.; Mitchell, T.K.; Dean, R.A. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 2007, 273, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S. Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol. 2012, 835, 255–269. [Google Scholar] [PubMed]
- Catlett, N.L.; Lee, B.N.; Yoder, O.C.; Turgeon, B.G. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 2003, 50, 9–11. [Google Scholar] [CrossRef]
- Goswami, R.S.; Xu, J.R.; Trail, F.; Hilburn, K.; Kistler, H.C. Genomic analysis of host-pathogen interaction between fusarium graminearum and wheat during early stages of disease development. Microbiology 2006, 152, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Phale, P.S.; Durani, S.; Wangikar, P.P. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 2003, 83, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. Swiss-model: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Ferraroni, M.; Myasoedova, N.M.; Schmatchenko, V.; Leontievsky, A.A.; Golovleva, L.A.; Scozzafava, A.; Briganti, F. Crystal structure of a blue laccase from lentinus tigrinus: Evidences for intermediates in the molecular oxygen reductive splitting by multicopper oxidases. BMC Struct. Biol. 2007, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Canters, G.W.; Gilardi, G. Engineering type 1 copper sites in proteins. FEBS Lett. 1993, 325, 39–48. [Google Scholar] [CrossRef]
- Daroch, M.; Houghton, C.A.; Moore, J.K.; Wilkinson, M.C.; Carnell, A.J.; Bates, A.D.; Iwanejko, L.A. Glycosylated yellow laccases of the basidiomycete stropharia aeruginosa. Enzyme Microb. Technol. 2014, 58–59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ge, W.; Buswell, J.A. Molecular cloning of a new laccase from the edible straw mushroom volvariella volvacea: Possible involvement in fruit body development. FEMS Microbiol. Lett. 2004, 230, 171–176. [Google Scholar] [CrossRef]
- Leonowicz, A.; Cho, N.S.; Luterek, J.; Wilkolazka, A.; Wojtas-Wasilewska, M.; Matuszewska, A.; Hofrichter, M.; Wesenberg, D.; Rogalski, J. Fungal laccase: Properties and activity on lignin. J. Basic Microbiol. 2001, 41, 185–227. [Google Scholar] [CrossRef]
- Piscitelli, A.; Pezzella, C.; Giardina, P.; Faraco, V.; Giovanni, S. Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 2010, 1, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Singh Arora, D.; Kumar Sharma, R. Ligninolytic fungal laccases and their biotechnological applications. Appl. Biochem. Biotechnol. 2010, 160, 1760–1788. [Google Scholar] [CrossRef] [PubMed]
- Rühl, M.; Fischer, C.; Kües, U. Ligninolytic enzyme activities alternate with mushroom production during industrial cultivation of pleurotus ostreatus on wheat straw-based substrate. Curr. Trends Biotechnol. Pharm. 2008, 2, 478–492. [Google Scholar]
- Isikhuemhen, O.S.; Mikiashvili, N.A.; Kelkar, V. Application of solid waste from anaerobic digestion of poultry litter in agrocybe aegerita cultivation: Mushroom production, lignocellulolytic enzymes activity and substrate utilization. Biodegradation 2009, 20, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Bonnen, A.M.; Anton, L.H.; Orth, A.B. Lignin-degrading enzymes of the commercial button mushroom, Agaricus bisporus. Appl. Environ. Microbiol. 1994, 60, 960–965. [Google Scholar] [PubMed]
- Langfelder, K.; Streibel, M.; Jahn, B.; Haase, G.; Brakhage, A.A. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 2003, 38, 143–158. [Google Scholar] [CrossRef]
- Fan, X.; Zhou, Y.; Xiao, Y.; Xu, Z.; Bian, Y. Cloning, expression and phylogenetic analysis of a divergent laccase multigene family in auricularia auricula-judae. Microbiol. Res. 2014, 169, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Matcham, S.E.; Jordan, B.R.; Wood, D.A. Estimation of fungal biomass in a solid substrate by three independent methods. Appl. Microbiol. Biotechnol. 1985, 21, 108–112. [Google Scholar] [CrossRef]
- Prillinger, H.; Esser, K. The phenoloxidases of the ascomycete podospora anserina. Xiii. Action and interaction of genes controlling the formation of laccase. Mol. Gen. Genet. 1977, 156, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Kwon, O.C.; Son, E.S.; Yoon, D.E.; Han, W.; Yoo, Y.B.; Lee, C.S. Taxonomy of Ganoderma lucidum from Korea based on rDNA and partial beta-tubulin gene sequence analysis. Mycobiology 2012, 40, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Ritter, G.J.; Seborg, R.M.; Mitchell, R.L. Factors affecting quantitative determination of lignin by 72 per cent sulfuric acid method. Ind. Eng. Chem. Anal. Ed. 1932, 4, 202–204. [Google Scholar] [CrossRef]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, W.; Li, J.; Feng, H.; You, S.; Zhang, L.; Norvienyeku, J.; Hu, K.; Sun, S.; Wang, Z. Importance of a Laccase Gene (Lcc1) in the Development of Ganoderma tsugae. Int. J. Mol. Sci. 2018, 19, 471. https://doi.org/10.3390/ijms19020471
Jin W, Li J, Feng H, You S, Zhang L, Norvienyeku J, Hu K, Sun S, Wang Z. Importance of a Laccase Gene (Lcc1) in the Development of Ganoderma tsugae. International Journal of Molecular Sciences. 2018; 19(2):471. https://doi.org/10.3390/ijms19020471
Chicago/Turabian StyleJin, Wensong, Jiahuan Li, Hongchang Feng, Si You, Liaoyuan Zhang, Justice Norvienyeku, Kaihui Hu, Shujing Sun, and Zonghua Wang. 2018. "Importance of a Laccase Gene (Lcc1) in the Development of Ganoderma tsugae" International Journal of Molecular Sciences 19, no. 2: 471. https://doi.org/10.3390/ijms19020471