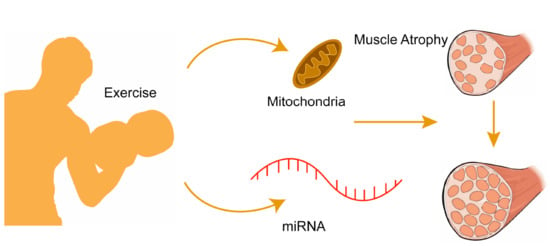
Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention
Abstract
1. Introduction
2. Functional Mechanism and Classification of miRNAs
3. MiRNAs for Regulating the Development of Skeletal Muscle
4. MiRNA and Muscle Atrophy
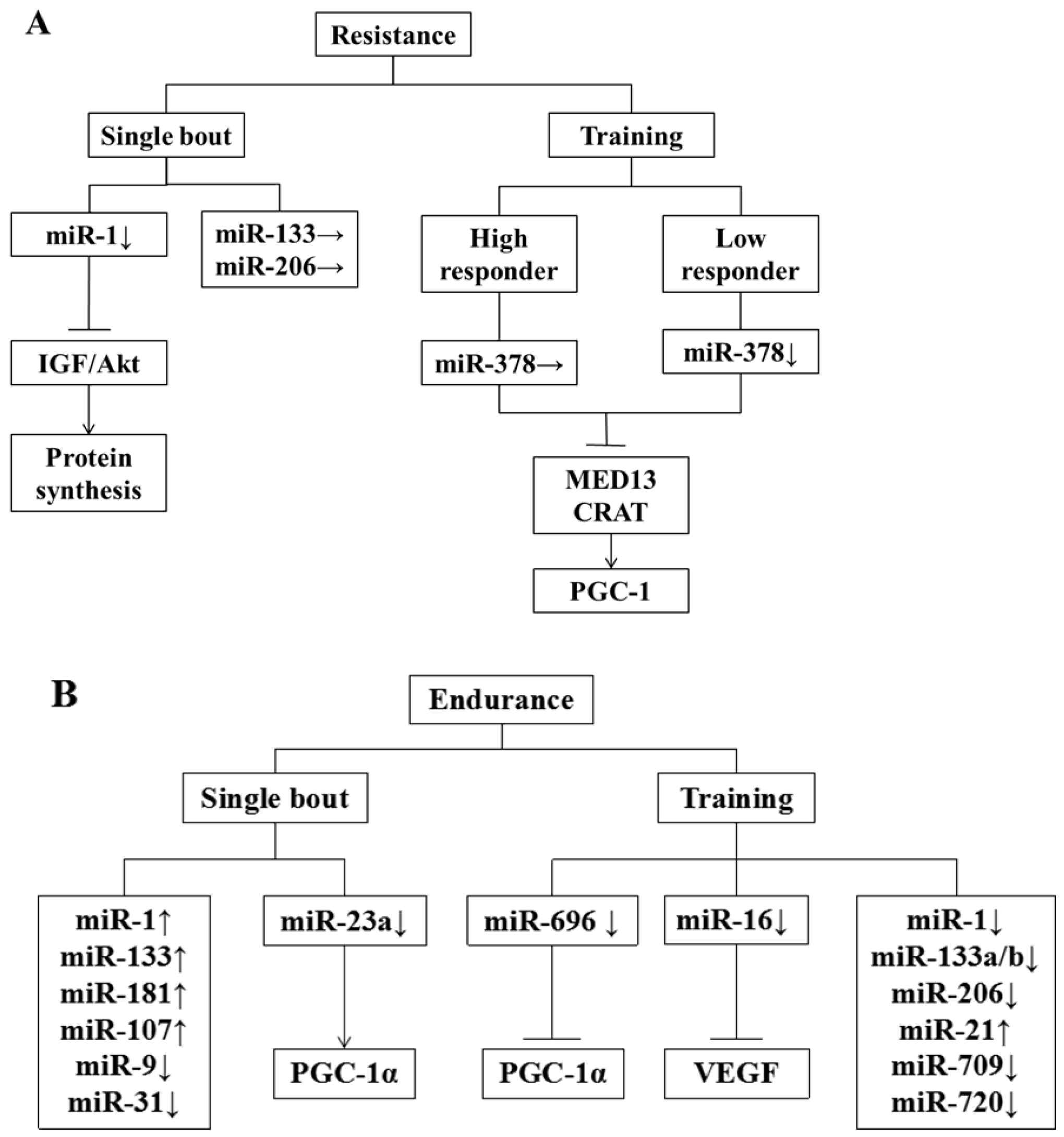
5. Effect of Exercise on miRNA
6. MiRNA as a Novel Research Direction of Muscle Atrophy
7. Conclusions and Expectation
- (a)
- The abnormal regulation of miRNAs can lead to a series of diseases such as DMD, ALS and sarcopenia. Whether miRNAs can be used as the new therapeutic targets or agents to detect and treat these diseases is highly required to be explored.
- (b)
- The changes in miRNAs and related proteins are observed in diverse muscle disorders and diseases. Whether miRNAs can be regulated in human body by generic and biochemical means is necessary to be explored, so as to accomplish the prevention and treatment of skeletal muscle injuries or corresponding disorders.
- (c)
- Exercise can modulate the expression of miRNAs for controlling a series of physiological and pathological changes. Exploring exercise-induced miRNAs as the targets will be beneficial for the development of novel and effective nutritional supplements, drugs or other intervention strategies for disease prevention and treatment or health promotion.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takemoto, Y.; Fukada, S.I. Molecular mechanism maintaining muscle satellite cells and the roles in sarcopenia. Clin. Calcium 2017, 27, 339–344. [Google Scholar] [PubMed]
- Bohnert, K.R.; McMillan, J.D.; Kumar, A. Emerging Roles of ER Stress and Unfolded Protein Response Pathways in Skeletal Muscle Health and Disease. J. Cell. Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Bombieri, F.; Schena, F.; Pellegrini, B.; Barone, P.; Tinazzi, M.; Erro, R. Walking on four limbs: A systematic review of Nordic Walking in Parkinson disease. Parkinsonism Relat. Disord. 2017, 38, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Kasukawa, Y.; Miyakoshi, N.; Hongo, M.; Ishikawa, Y.; Kudo, D.; Suzuki, M.; Mizutani, T.; Kimura, R.; Ono, Y.; Shimada, Y. Age-related changes in muscle strength and spinal kyphosis angles in an elderly Japanese population. Clin. Interv. Aging 2017, 12, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Kou, X.; Yang, Y.; Chen, N. MicroRNA-Regulated Proinflammatory Cytokines in Sarcopenia. Mediat. Inflamm. 2016, 2016, 1438686. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Sambandan, S.; Akbalik, G.; Kochen, L.; Rinne, J.; Kahlstatt, J.; Glock, C.; Tushev, G.; Alvarez-Castelao, B.; Heckel, A.; Schuman, E.M. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 2017, 355, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Dennis, C.; Brasset, E.; Sarkar, A.; Vaury, C. Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila. Nat. Commun. 2016, 7, 13739. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Xi, Y. MicroRNAs mediate therapeutic and preventive effects of natural agents in breast cancer. Chin. J. Nat. Med. 2016, 14, 881–887. [Google Scholar] [CrossRef]
- McCarthy, J.J. The MyomiR network in skeletal muscle plasticity. Exerc. Sport Sci. Rev. 2011, 39, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. USA 2010, 107, 4218–4223. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.J.; Chaillou, T.; McCarthy, J.J. The role of microRNAs in skeletal muscle health and disease. Front. Biosci. 2015, 20, 37–77. [Google Scholar]
- Buckingham, M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Nie, M.; Deng, Z.L.; Liu, J.; Wang, D.Z. Noncoding RNAs, Emerging Regulators of Skeletal Muscle Development and Diseases. BioMed Res. Int. 2015, 2015, 676575. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Chen, J. MicroRNAs in skeletal myogenesis. Cell Cycle 2011, 10, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Potthoff, M.J.; Olson, E.N. MEF2: A central regulator of diverse developmental programs. Development 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed]
- Harding, R.L.; Velleman, S.G. MicroRNA regulation of myogenic satellite cell proliferation and differentiation. Mol. Cell. Biochem. 2016, 412, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, Y.; Tanihata, J.; Komaki, H.; Ishiyama, A.; Oya, Y.; Ruegg, U.; Takeda, S.I.; Hashido, K. Characterization and Functional Analysis of Extracellular Vesicles and Muscle-Abundant miRNAs (miR-1, miR-133a, and miR-206) in C2C12 Myocytes and mdx Mice. PLoS ONE 2016, 11, e0167811. [Google Scholar] [CrossRef] [PubMed]
- Pollock, R.; Treisman, R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991, 5, 2327–2341. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Lee, Y.S.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ge, Y.; Drnevich, J.; Zhao, Y.; Band, M.; Chen, J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 2010, 189, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.; Catoe, H.; Werner, R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006, 34, 5863–5871. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xie, S.; Luan, J.; Zhou, X.; Han, J. Quantitative proteomics and protein network analysis of A549 lung cancer cells affected by miR-206. Biosci. Trends 2013, 7, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Li, R.C.; Tao, J.; Guo, Y.B.; Wu, H.D.; Liu, R.F.; Bai, Y.; Lv, Z.Z.; Luo, G.Z.; Li, L.L.; Wang, M.; et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ. Res. 2013, 112, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, Y.; Yang, G.; Chen, X.; Zhang, Y.; Cao, G.; Wang, J.; Sun, Y.; Zhang, P.; Fan, M.; et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008, 36, 2690–2699. [Google Scholar] [CrossRef] [PubMed]
- Bakre, A.; Wu, W.; Hiscox, J.; Spann, K.; Teng, M.N.; Tripp, R.A. Human respiratory syncytial virus non-structural protein NS1 modifies miR-24 expression via transforming growth factor-beta. J. Gen. Virol. 2015, 96, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Crist, C.G.; Montarras, D.; Pallafacchina, G.; Rocancourt, D.; Cumano, A.; Conway, S.J.; Buckingham, M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. USA 2009, 106, 13383–13387. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kuroiwa, A. Hoxa-11 and Hoxa-13 are involved in repression of MyoD during limb muscle development. Dev. Growth Differ. 2003, 45, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, B.; Castellani, L.; Fasanaro, P.; Basso, A.; Alema, S.; Martelli, F.; Falcone, G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE 2009, 4, e7607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Sun, Y.; Chen, J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011, 192, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Zheng, Y.; Li, F.; Lu, Z.; Chen, C.; Liu, J.; Wang, Y.; Peng, Y.; Shen, Z.; et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp. Cell Res. 2012, 318, 2324–2334. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, Y.F.; Wu, G.F.; Song, Z.Y.; Lu, H.Z.; Song, C.C.; Zhang, Q.L.; Zhu, J.Y.; Yang, G.S.; Shi, X.E. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int. J. Mol. Sci. 2013, 15, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.; Mastroyiannopoulos, N.P.; Uney, J.B.; Phylactou, L.A. miR-186 inhibits muscle cell differentiation through myogenin regulation. J. Biol. Chem. 2014, 289, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H. MicroRNA in myogenesis and muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Schuenke, M.D.; Reed, D.W.; Kraemer, W.J.; Staron, R.S.; Volek, J.S.; Hymer, W.C.; Gordon, S.; Perry Koziris, L. Effects of 14 days of microgravity on fast hindlimb and diaphragm muscles of the rat. Eur. J. Appl. Physiol. 2009, 106, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.L.; Bandstra, E.R.; Harrison, B.C.; Thorng, S.; Stodieck, L.S.; Kostenuik, P.J.; Morony, S.; Lacey, D.L.; Hammond, T.G.; Leinwand, L.L.; et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 2009, 106, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Enoki, Y.; Watanabe, H.; Arake, R.; Sugimoto, R.; Imafuku, T.; Tominaga, Y.; Ishima, Y.; Kotani, S.; Nakajima, M.; Tanaka, M.; et al. Indoxyl sulfate potentiates skeletal muscle atrophy by inducing the oxidative stress-mediated expression of myostatin and atrogin-1. Sci. Rep. 2016, 6, 32084. [Google Scholar] [CrossRef] [PubMed]
- Leijs, M.M.; Koppe, J.G.; Olie, K.; van Aalderen, W.M.; de Voogt, P.; ten Tusscher, G.W. Effects of dioxins, PCBs, and PBDEs on immunology and hematology in adolescents. Environ. Sci. Technol. 2009, 43, 7946–7951. [Google Scholar] [CrossRef] [PubMed]
- Ringholm, S.; Bienso, R.S.; Kiilerich, K.; Guadalupe-Grau, A.; Aachmann-Andersen, N.J.; Saltin, B.; Plomgaard, P.; Lundby, C.; Wojtaszewski, J.F.; Calbet, J.A.; et al. Bed rest reduces metabolic protein content and abolishes exercise-induced mRNA responses in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E649–E658. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, Y.; Takeda, K.; Tamura, Y.; Fujimaki, S.; Takemasa, T.; Hatta, H. Nrf2 deficiency does not affect denervation-induced alterations in mitochondrial fission and fusion proteins in skeletal muscle. Physiol. Rep. 2016, 4, e13064. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; McCarthy, J.J.; Fry, C.S.; Esser, K.A.; Rasmussen, B.B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1333–E1340. [Google Scholar] [CrossRef] [PubMed]
- Hamrick, M.W.; Herberg, S.; Arounleut, P.; He, H.Z.; Shiver, A.; Qi, R.Q.; Zhou, L.; Isales, C.M.; Mi, Q.S. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem. Biophys. Res. Commun. 2010, 400, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Wang, Y.; Li, Y.; Cui, L.; Zhao, Y.; Zhao, B.; Li, K. MiR-206, a key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. 2015, 11, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Walden, T.B.; Timmons, J.A.; Keller, P.; Nedergaard, J.; Cannon, B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J. Cell. Physiol. 2009, 218, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Mercken, E.M.; Majounie, E.; Ding, J.; Guo, R.; Kim, J.; Bernier, M.; Mattison, J.; Cookson, M.R.; Gorospe, M.; de Cabo, R.; et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging 2013, 5, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, E.R.; Kawamoto, E.M.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H., III; Lehrmann, E.; Camandola, S.; Becker, K.G.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Kong, M.; Ye, Y.; Hong, S.; Cheng, L.; Jiang, L. Serum miR-206 and other muscle-specific microRNAs as non-invasive biomarkers for Duchenne muscular dystrophy. J. Neurochem. 2014, 129, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.S.; Kessler, C.B.; Shenoy, V.; Rosen, C.J.; Delany, A.M. IGF-I 3′ untranslated region: Strain-specific polymorphisms and motifs regulating IGF-I in osteoblasts. Endocrinology 2013, 154, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Iwama, H.; Deguchi, K.; Ikeda, K.; Takata, T.; Kokudo, Y.; Kamada, M.; Fujikawa, K.; Hirose, K.; Masugata, H.; et al. Serum microRNA expression profiling in patients with multiple system atrophy. Mol. Med. Rep. 2018, 17, 852–860. [Google Scholar] [PubMed]
- Koutsoulidou, A.; Photiades, M.; Kyriakides, T.C.; Georgiou, K.; Prokopi, M.; Kapnisis, K.; Lusakowska, A.; Nearchou, M.; Christou, Y.; Papadimas, G.K.; et al. Identification of exosomal muscle-specific miRNAs in serum of myotonic dystrophy patients relating to muscle disease progress. Hum. Mol. Genet. 2017, 26, 3285–3302. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Lee, K.P.; Milholland, B.; Shin, Y.J.; Kang, J.S.; Kwon, K.S.; Suh, Y. Comprehensive miRNA Profiling of Skeletal Muscle and Serum in Induced and Normal Mouse Muscle Atrophy During Aging. J. Gerontol. 2017, 72, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Nakamura, A.; Aoki, Y.; Ito, N.; Kishi, S.; Yamamoto, K.; Sekiguchi, M.; Takeda, S.; Hashido, K. Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: Promising novel blood-based markers for muscular dystrophy. PLoS ONE 2011, 6, e18388. [Google Scholar] [CrossRef] [PubMed]
- Cacchiarelli, D.; Incitti, T.; Martone, J.; Cesana, M.; Cazzella, V.; Santini, T.; Sthandier, O.; Bozzoni, I. miR-31 modulates dystrophin expression: New implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011, 12, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Signaling in muscle atrophy and hypertrophy. Physiology 2008, 23, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.A.; Alessi, D.R.; Cohen, P.; Andjelkovich, M.; Hemmings, B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995, 378, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Elia, L.; Contu, R.; Quintavalle, M.; Varrone, F.; Chimenti, C.; Russo, M.A.; Cimino, V.; De Marinis, L.; Frustaci, A.; Catalucci, D.; et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 2009, 120, 2377–2385. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Zhang, Y.; Ren, J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: Role of microRNA-1 and microRNA-133a. J. Cell. Mol. Med. 2012, 16, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhu, C.D.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 2013, 216, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Juan, H.C.; Tsai, H.T.; Chang, P.H.; Huang, C.Y.; Hu, C.P.; Wong, F.H. Insulin-like growth factor 1 mediates 5-fluorouracil chemoresistance in esophageal carcinoma cells through increasing survivin stability. Apoptosis 2011, 16, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, R.; Workeneh, B.; Dong, Y.; Wang, X.; Hu, Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012, 82, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Dutta, A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Sci. 2013, 126, 2678–2691. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Baek, M.; Gusev, Y.; Brackett, D.J.; Nuovo, G.J.; Schmittgen, T.D. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 2008, 14, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, N.; Alexander, M.S.; Casar, J.C.; Kunkel, L.M. Identification of a novel microRNA that regulates the proliferation and differentiation in muscle side population cells. Stem Cells Dev. 2012, 21, 3031–3043. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.M.; Wang, J.; Yan, Z.; You, Y.P.; Li, C.Y.; Qian, X.; Yin, Y.; Zhao, P.; Wang, Y.Y.; Wang, X.F.; et al. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 2012, 7, e32709. [Google Scholar] [CrossRef] [PubMed]
- Rebbapragada, A.; Benchabane, H.; Wrana, J.L.; Celeste, A.J.; Attisano, L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 2003, 23, 7230–7242. [Google Scholar] [CrossRef] [PubMed]
- McCroskery, S.; Thomas, M.; Maxwell, L.; Sharma, M.; Kambadur, R. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 2003, 162, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Welle, S.; Bhatt, K.; Pinkert, C.A. Myofibrillar protein synthesis in myostatin-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E409–E415. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Huynh, T.V.; Lee, Y.S.; Sebald, S.M.; Wilcox-Adelman, S.A.; Iwamori, N.; Lepper, C.; Matzuk, M.M.; Fan, C.M. Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc. Natl. Acad. Sci. USA 2012, 109, E2353–E2360. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; McPherron, A.C. Myostatin inhibition induces muscle fibre hypertrophy prior to satellite cell activation. J. Physiol. 2012, 590, 2151–2165. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, X.; Yu, B.; He, J.; Chen, D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem. Biophys. Res. Commun. 2012, 423, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.L.; Loh, A.S. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 2011, 300, C124–C137. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, X.; Chen, D. Myostatin: A novel insight into its role in metabolism, signal pathways, and expression regulation. Cell. Signal. 2011, 23, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gonzalo, R.; Fernandez-Gonzalo, S.; Turon, M.; Prieto, C.; Tesch, P.A.; Garcia-Carreira Mdel, C. Muscle, functional and cognitive adaptations after flywheel resistance training in stroke patients: A pilot randomized controlled trial. J. Neuroeng. Rehabilit. 2016, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Junior, R.M.; Berton, R.; de Souza, T.M.; Chacon-Mikahil, M.P.; Cavaglieri, C.R. Effect of the flexibility training performed immediately before resistance training on muscle hypertrophy, maximum strength and flexibility. Eur. J. Appl. Physiol. 2017, 117, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Vitucci, D.; Labruna, G.; Imperlini, E.; Randers, M.B.; Schmidt, J.F.; Hagman, M.; Andersen, T.R.; Russo, R.; Orru, S.; et al. Effect of lifelong football training on the expression of muscle molecular markers involved in healthy longevity. Eur. J. Appl. Physiol. 2017, 117, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Lew, J.K.; Pearson, J.T.; Schwenke, D.O.; Katare, R. Exercise mediated protection of diabetic heart through modulation of microRNA mediated molecular pathways. Cardiovasc. Diabetol. 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Kobayashi, R.; Sakamaki-Sunaga, M. Effect of Resistance Exercise on Arterial Stiffness during the Follicular and Luteal Phases of the Menstrual Cycle. Int. J. Sports Med. 2017, 38, 347. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; Glynn, E.L.; Fry, C.S.; Dhanani, S.; Volpi, E.; Rasmussen, B.B. Essential amino acids increase microRNA-499, -208b, and -23a and downregulate myostatin and myocyte enhancer factor 2C mRNA expression in human skeletal muscle. J. Nutr. 2009, 139, 2279–2284. [Google Scholar] [CrossRef] [PubMed]
- Matheny, R.W., Jr.; Carrigan, C.T.; Abdalla, M.N.; Geddis, A.V.; Leandry, L.A.; Aguilar, C.A.; Hobbs, S.S.; Urso, M.L. RNA transcript expression of IGF-I/PI3K pathway components in regenerating skeletal muscle is sensitive to initial injury intensity. Growth Horm. IGF Res. 2017, 32, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Davidsen, P.K.; Gallagher, I.J.; Hartman, J.W.; Tarnopolsky, M.A.; Dela, F.; Helge, J.W.; Timmons, J.A.; Phillips, S.M. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 2011, 110, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J. Biol. Chem. 2011, 286, 19431–19438. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.J.; McCarthy, J.J. MicroRNAs in skeletal muscle biology and exercise adaptation. Free Radic. Biol. Med. 2013, 64, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Scheele, C.; Yfanti, C.; Akerstrom, T.; Nielsen, A.R.; Pedersen, B.K.; Laye, M.J. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010, 588, 4029–4037. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, D.; Goljanek, K.; Rathjen, T.; Oustanina, S.; Braun, T.; Dalmay, T.; Munsterberg, A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008, 321, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Guller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Abadi, A.; Akhtar, M.; Hettinga, B.P.; Tarnopolsky, M.A. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE 2009, 4, e5610. [Google Scholar] [CrossRef] [PubMed]
- Aoi, W.; Naito, Y.; Mizushima, K.; Takanami, Y.; Kawai, Y.; Ichikawa, H.; Yoshikawa, T. The microRNA miR-696 regulates PGC-1α in mouse skeletal muscle in response to physical activity. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E799–E806. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Li, P.; Jia, W.; Jiang, T.; Wang, Z.; Xiang, Y. miR-696 plays a role in hepatic gluconeogenesis in ob/ob mice by targeting PGC-1α. Int. J. Mol. Med. 2016, 38, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Pereira, R.I.; Erickson, C.B.; Swibas, T.A.; Kang, C.; Van Pelt, R.E. Time since menopause and skeletal muscle estrogen receptors, PGC-1α, and AMPK. Menopause 2017, 24, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.J.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589, 3983–3994. [Google Scholar] [CrossRef] [PubMed]
- Aoi, W.; Ichikawa, H.; Mune, K.; Tanimura, Y.; Mizushima, K.; Naito, Y.; Yoshikawa, T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Sawada, S.; Kon, M.; Wada, S.; Ushida, T.; Suzuki, K.; Akimoto, T. Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PLoS ONE 2013, 8, e70823. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.S.; Kawahara, G.; Motohashi, N.; Casar, J.C.; Eisenberg, I.; Myers, J.A.; Gasperini, M.J.; Estrella, E.A.; Kho, A.T.; Mitsuhashi, S.; et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 2013, 20, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Snyder, C.M.; Rice, A.L.; Estrella, N.L.; Held, A.; Kandarian, S.C.; Naya, F.J. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development 2013, 140, 31–42. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Chen, N. Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention. Int. J. Mol. Sci. 2018, 19, 405. https://doi.org/10.3390/ijms19020405
Zhang S, Chen N. Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention. International Journal of Molecular Sciences. 2018; 19(2):405. https://doi.org/10.3390/ijms19020405
Chicago/Turabian StyleZhang, Shufang, and Ning Chen. 2018. "Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention" International Journal of Molecular Sciences 19, no. 2: 405. https://doi.org/10.3390/ijms19020405
APA StyleZhang, S., & Chen, N. (2018). Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention. International Journal of Molecular Sciences, 19(2), 405. https://doi.org/10.3390/ijms19020405