Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites
Abstract
:1. Introduction
2. Telomerase Origin
3. Telomerase Architecture and Biogenesis
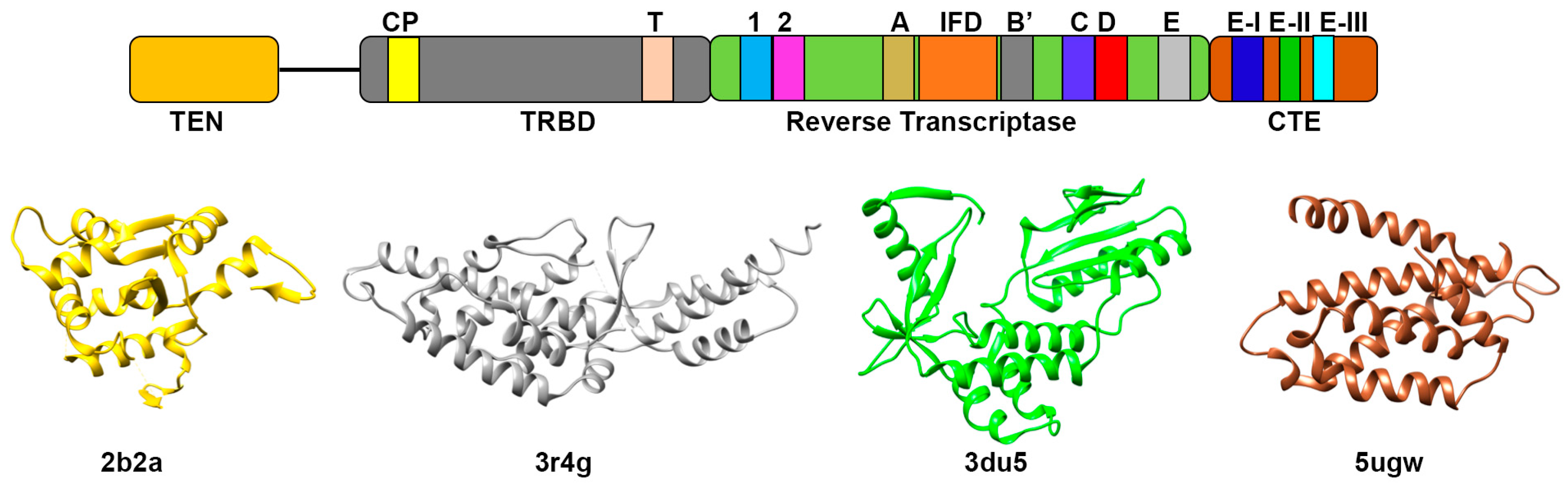
3.1. Telomerase Reverse Transcriptase Protein (TERT)
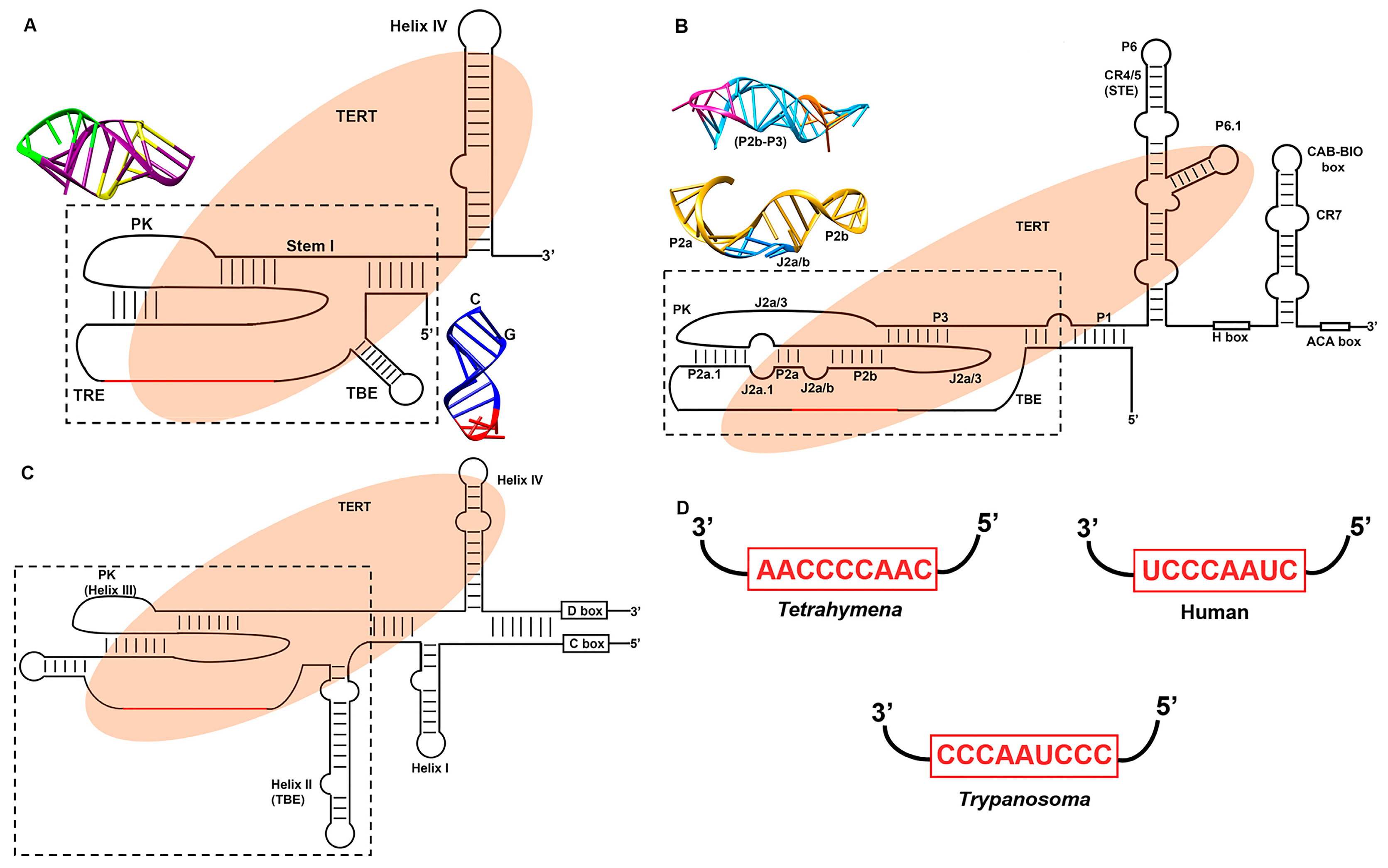
3.2. Telomerase RNA (TER)
3.2.1. The Template
3.2.2. Template Boundary Element (TBE)
3.2.3. Pseudoknot
3.2.4. Stem Terminus Element (STE)
3.3. Structural Variation in Telomerase RNA
3.3.1. Ciliate Telomerase RNA Structure
3.3.2. Human Telomerase RNA Structure
3.3.3. Flagellate Telomerase RNA Structure
3.4. Telomerase Biogenesis and Maturation
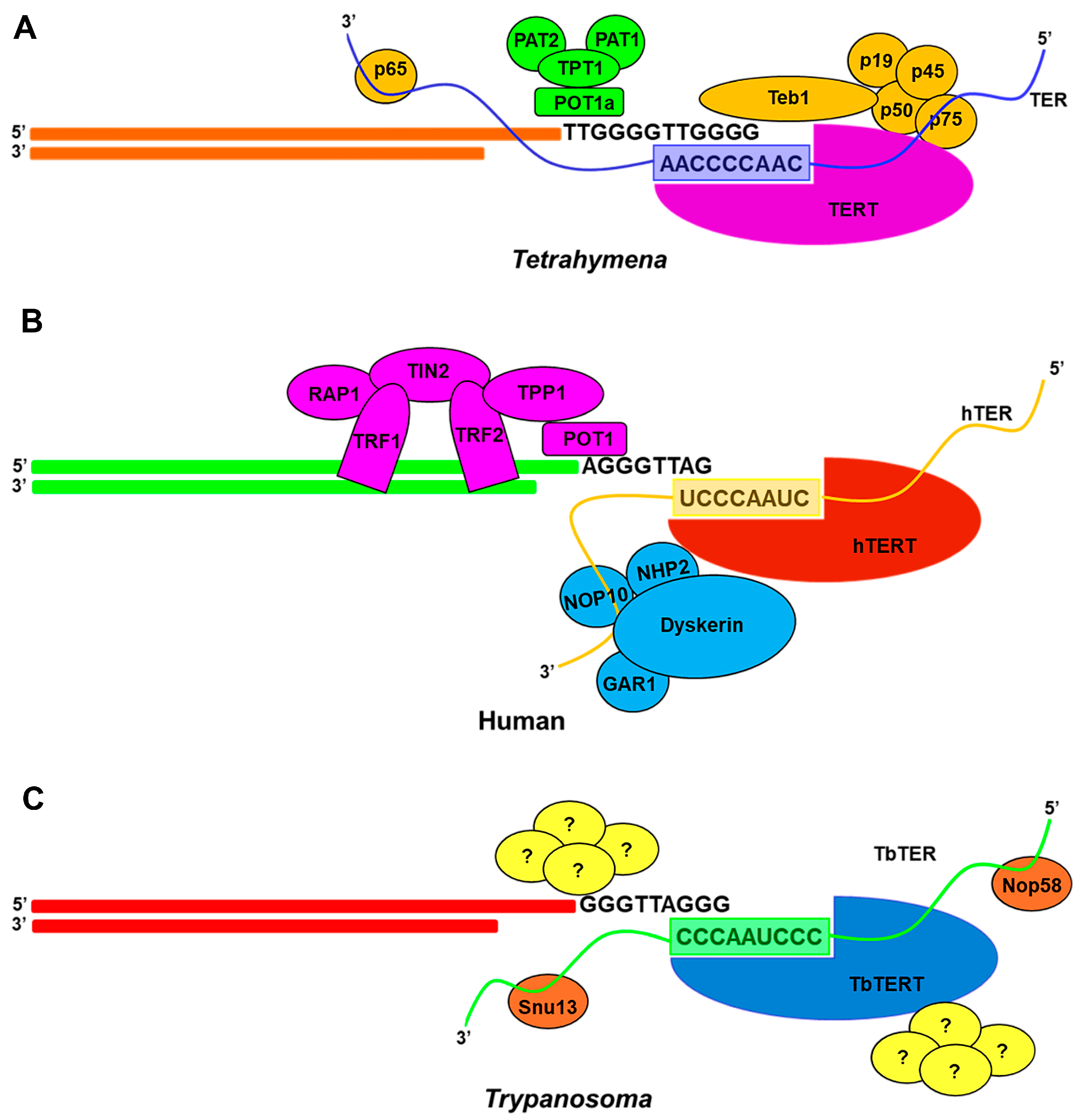
3.4.1. Ciliate Telomerase RNP Maturation
3.4.2. Yeast Telomerase RNP Maturation
3.4.3. Human Telomerase RNP Maturation
3.4.4. Flagellates Telomerase RNP Maturation
4. Recruitment of Telomerase RNPs to Telomere
4.1. Coupling of Ciliate RNP to Telomere
4.2. Coupling of Yeast RNP to Telomere
4.3. Coupling of Human RNP to Telomere
4.4. Coupling of Flagellates RNP to Telomere
5. Factors Involved in Telomerase Assembly and Activity
6. Pathophysiology of Telomerase
6.1. Ageing, Cellular Immortality and Cancer
6.2. Dyskeratosis Congenita: Case of Telomerase Dysfunction
6.3. Telomere, Telomerase and Human Pathogens
6.3.1. Telomere, Telomerase and Virulence in Trypanosoma brucei
6.3.2. Telomere, Telomerase and Virulence in Plasmodium falciparum
7. Conclusions and Future Direction
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RNP | Ribonucleoprotein |
| TER | Telomerase RNA |
| TERT | Telomerase Reverse Transcriptase |
| NHEJ | Non-Homologous End Joining |
| RDR | Recombination-Dependent Replication |
| PLE | Penelope Like Elements |
| ALT | Alternative Lengthening of Telomerase |
| TEN | Telomerase “Essential” N-terminal |
| TRBD | Telomerase Binding Domain |
| RT | Reverse Transcriptase |
| CTE | C-terminal Extension |
| RAP | Repeat Addition Processivity |
| TBE | Template Boundary Element |
| PK | Pseudoknot |
| STE | Stem Terminus Element |
| CR4/5 | Conserved Region4/5 |
| snRNA | Small Nuclear RNA |
| snoRNA | Small Nucleolar RNA |
| CAB | Cajal Body |
| scaRNA | Small Cajal Body RNA |
| TASC | Telomere Adaptor Sub-Complex |
| RPA | Replication Protein A |
| VSG | Variable Surface Glycoprotein |
| GC | Gene Conversion |
| TE | Telomeric Exchange |
| DSB | Double Stranded Break |
| TAS | Telomere Associated Sequences |
| TAREs | Telomere Associated Repeat Elements |
| TPE | Telomere Position Effect |
References
- De Lange, T. A loopy view of telomere evolution. Front. Genet. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D. Origin of concatameric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.L.; Bradley, J.D.; Attardi, L.D.; Blackburn, E.H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 1990, 344, 126–132. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. How telomeres solve the end-protection problem. Science 2009, 326, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; DeBaryshe, P.G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003, 37, 485–511. [Google Scholar] [CrossRef] [PubMed]
- Salanti, A.; Clausen, T.M.; Agerbaek, M.O.; Al Nakouzi, N.; Dahlback, M.; Oo, H.Z.; Lee, S.; Gustavsson, T.; Rich, J.R.; Hedberg, B.J.; et al. Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein. Cancer Cell 2015, 28, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Hovel-Miner, G.A.; Boothroyd, C.E.; Mugnier, M.; Dreesen, O.; Cross, G.A.; Papavasiliou, F.N. Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog. 2012, 8, e1002900. [Google Scholar] [CrossRef] [PubMed]
- Jaskelioff, M.; Muller, F.L.; Paik, J.H.; Thomas, E.; Jiang, S.; Adams, A.C.; Sahin, E.; Kost-Alimova, M.; Protopopov, A.; Cadinanos, J.; et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 2011, 469, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Dokal, I. Dyskeratosis congenita. Hematol. Am. Soc. Hematol. Educ. Program. 2011, 2011, 480–486. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004, 5, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.M.; Morin, G.B.; Chapman, K.B.; Weinrich, S.L.; Andrews, W.H.; Lingner, J.; Harley, C.B.; Cech, T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Boeke, J.D. The unusual phylogenetic distribution of retrotransposons: A hypothesis. Genome Res. 2003, 13, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Dreesen, O.; Li, B.; Cross, G.A. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acid Res. 2005, 33, 4536–4543. [Google Scholar] [CrossRef] [PubMed]
- Giardini, M.A.; Lira, C.B.; Conte, F.F.; Camillo, L.R.; de Siqueira Neto, J.L.; Ramos, C.H.; Cano, M.I. The putative telomerase reverse transcriptase component of Leishmania amazonensis: Gene cloning and characterization. Parasitol. Res. 2006, 98, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; Danilevskaya, O.N.; Traverse, K.L.; Lowenhaupt, K. Evolutionary links between telomeres and transposable elements. Genetica 1997, 100, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, T.H. Telomerase and retrotransposons: Which came first? Science 1997, 277, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.A.; Arkhipova, I.R. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 9352–9357. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Kajikawa, M.; Matsumoto, T.; Okada, N. Mechanism by which a LINE protein recognizes its 3′ tail RNA. Nucleic Acid Res 2014, 42, 10605–10617. [Google Scholar] [CrossRef] [PubMed]
- Jamburuthugoda, V.K.; Eickbush, T.H. Identification of RNA binding motifs in the R2 retrotransposon-encoded reverse transcriptase. Nucleic Acid Res. 2014, 42, 8405–8415. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, Y.; Honda, S.; Hoffmann, S.; Marz, M.; Mosig, A.; Podlevsky, J.D.; Stadler, P.F.; Selker, E.U.; Chen, J.J. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acid Res. 2013, 41, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Sperger, J.M.; Chapman, K.B.; Cech, T.R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. USA 1998, 95, 8479–8484. [Google Scholar] [CrossRef] [PubMed]
- Erwin, J.A.; Marchetto, M.C.; Gage, F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014, 8, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.D.; Neumann, A.A.; Yeager, T.R.; Reddel, R.R. Alternative lengthening of telomeres in mammalian cells. Oncogene 2002, 21, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Lendvay, T.S.; Morris, D.K.; Sah, J.; Balasubramanian, B.; Lundblad, V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 1996, 144, 1399–1412. [Google Scholar] [PubMed]
- Lingner, J.; Cech, T.R. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. USA 1996, 193, 10712–10717. [Google Scholar] [CrossRef]
- Collins, K.; Gandhi, L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 1998, 95, 8485–8590. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.; Zhou, W.; McPhail, T.; Oulton, R.; Yeung, D.S.; Mar, V.; Bass, M.B.; Robinson, M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997, 11, 3109–3115. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.A.; Allsopp, R.C.; Chin, L.; Morin, G.B.; DePinho, R.A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 1998, 16, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.M.; Rocha, E.P.; Mancio-Silva, L.; Prevost, C.; Hernandez-Verdun, D.; Scherf, A. The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus. Nucleic Acids Res. 2005, 33, 11111–11122. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Lai, C.K.; Collins, K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005, 280, 17533–17539. [Google Scholar] [CrossRef] [PubMed]
- Robart, A.R.; Collins, K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell. 2011, 42, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Collins, K. Single-stranded DNA repeat synthesis by telomerase. Curr. Opin. Chem. Biol. 2011, 15, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.A.; Podell, E.R.; Cech, T.R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2017, 13, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Rouda, S.; Skordalakes, E. Structure of the RNA-binding domain of telomerase: Implications for RNA recognition and binding. Structure 2007, 15, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Lue, N.F.; Lin, Y.C.; Mian, I.S. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 2003, 23, 8440–8449. [Google Scholar] [CrossRef] [PubMed]
- Cote, M.L.; Roth, M.J. Murine leukemia virus reverse transcriptase: Structural comparison with HIV-1 reverse transcriptase. Virus Res. 2008, 134, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Gillis, A.J.; Schuller, A.P.; Skordalakes, E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 2008, 455, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Curcio, M.J.; Lue, N.F. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 20304–20310. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.; Gillis, A.; Futahashi, M.; Fujiwara, H.; Skordalakes, E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010, 17, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, H.; Rice, C.; Skordalakes, E. Structural Analysis Reveals the Deleterious Effects of Telomerase Mutations in Bone Marrow Failure Syndromes. J. Biol. Chem. 2017, 292, 4593–4601. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.J.; Romero, D.P. Phylogenetic relationships amongst tetrahymenine ciliates inferred by a comparison of telomerase RNAs. Int. J. Syst. Evol. Microbiol. 2002, 52, 2297–22302. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Mosig, A.; Qi, X.; Li, Y.; Stadler, P.F.; Chen, J.J. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J. Biol. Chem. 2008, 283, 2049–2059. [Google Scholar] [CrossRef] [PubMed]
- Gunisova, S.; Elboher, E.; Nosek, J.; Gorkovoy, V.; Brown, Y.; Lucier, J.F.; Laterreur, N.; Wellinger, R.J.; Tzfati, Y.; Tomaska, L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 2009, 15, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Sanford, S.; Basu, S.; Park, M.; Pandya, U.M.; Li, B.; Chakrabarti, K. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013, 23, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Kolet, L.; Doniger, T.; Biswas, V.K.; Unger, R.; Tzfati, Y.; Michaeli, S. The Trypanosoma brucei telomerase RNA (TER) homologue binds core proteins of the C/D snoRNA family. FEBS Lett. 2013, 587, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, K.; Pearson, M.; Grate, L.; Sterne-Weiler, T.; Deans, J.; Donohue, J.P.; Ares, M., Jr. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA 2007, 13, 1923–1939. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W. Telomerase is processive. Mol. Cell. Biol. 1991, 11, 4572–4580. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Arneric, M.; Lingner, J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007, 21, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.S.; Phillips, J.A.; Chan, A.; Sabourin, M.; Paeschke, K.; Zakian, V.A. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 2010, 17, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Greider, C.W. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003, 22, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Gilley, D.; Blackburn, E.H. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol. 1996, 16, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Smith, D.L.; Blackburn, E.H. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol. Biol. Cell 2004, 15, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Drosopoulos, W.C.; Direnzo, R.; Prasad, V.R. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 2005, 280, 32801–32810. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.F.; Podlevsky, J.D.; Qi, X.; Chen, Y.; Xie, M.; Chen, J.J. A self-regulating template in human telomerase. Proc. Natl. Acad. Sci. USA 2014, 111, 11311–11316. [Google Scholar] [CrossRef] [PubMed]
- Stohr, B.A.; Xu, L.; Blackburn, E.H. The terminal telomeric DNA sequence determines the mechanism of dysfunctional telomere fusion. Mol. Cell 2010, 39, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Chakrabarti, K. Structural definition of telomerase RNA in Trypanosoma brucei. 2018; manuscript in preparation. [Google Scholar]
- Autexier, C.; Greider, C.W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995, 9, 2227–2239. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Gomez, A.; Stone, M.D. A conserved motif in Tetrahymena thermophila telomerase reverse transcriptase is proximal to the RNA template and is essential for boundary definition. J. Biol. Chem. 2013, 288, 22141–22149. [Google Scholar] [CrossRef] [PubMed]
- Podlevsky, J.D.; Li, Y.; Chen, J.J. The functional requirement of two structural domains within telomerase RNA emerged early in eukaryotes. Nucleic Acid Res. 2016, 44, 9891–9901. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Greider, C.W. Template boundary definition in mammalian telomerase. Genes Dev. 2003, 17, 2747–2752. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, C.S.; Blasco, M.A.; Funk, W.D.; Feng, J.; Villeponteau, B.; Greider, C.W.; Herr, W. The mouse telomerase RNA 5′-end lies just upstream of the telomerase template sequence. Nucleic Acid Res. 1998, 26, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Blasco, M.A.; Greider, C.W. Secondary structure of vertebrate telomerase RNA. Cell 2000, 100, 503–514. [Google Scholar] [CrossRef]
- Tzfati, Y.; Knight, Z.; Roy, J.; Blackburn, E.H. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 2003, 17, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Cash, D.D.; Cohen-Zontag, O.; Kim, N.K.; Shefer, K.; Brown, Y.; Ulyanov, N.B.; Tzfati, Y.; Feigon, J. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 10970–10975. [Google Scholar] [CrossRef] [PubMed]
- Ly, H.; Blackburn, E.H.; Parslow, T.G. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol. Cell. Biol. 2003, 23, 6849–6856. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xie, M.; Brown, A.F.; Bley, C.J.; Podlevsky, J.D.; Chen, J.J. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 2012, 31, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Theimer, C.A.; Blois, C.A.; Feigon, J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 2005, 17, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Cech, T.R. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 2008, 15, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Kim, N.K.; Peterson, R.D.; Wang, Z.; Feigon, J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 18761–18768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Kim, N.K.; Feigon, J. Architecture of human telomerase RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20325–20332. [Google Scholar] [CrossRef] [PubMed]
- Mason, D.X.; Goneska, E.; Greider, C.W. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 2003, 23, 5606–5613. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Opperman, K.K.; Greider, C.W. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acid Res. 2002, 30, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Brown, Y.; Abraham, M.; Pearl, S.; Kabaha, M.M.; Elboher, E.; Tzfati, Y. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007, 35, 6280–6289. [Google Scholar] [CrossRef] [PubMed]
- McCormick-Graham, M.; Romero, D.P. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995, 23, 1091–1097. [Google Scholar] [CrossRef]
- Huang, J.; Brown, A.F.; Wu, J.; Xue, J.; Bley, C.J.; Rand, D.P.; Wu, L.; Zhang, R.; Chen, J.J.; Lei, M. Structural basis for protein-RNA recognition in telomerase. Nat. Struct. Mol. Biol. 2014, 21, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.D.; Mihalusova, M.; O’Connor, C.M.; Prathapam, R.; Collins, K.; Zhuang, X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 2007, 446, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.R.; Collins, K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 2000, 6, 361–371. [Google Scholar] [CrossRef]
- Lai, C.K.; Miller, M.C.; Collins, K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 2003, 11, 1673–1683. [Google Scholar] [CrossRef]
- Bley, C.J.; Qi, X.; Rand, D.P.; Borges, C.R.; Nelson, R.W.; Chen, J.J. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 20333–20338. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Collins, K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb. Prespect. Biol. 2011, 3, a003558. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.J.; Zakian, V.A. Telomerase RNA stem terminus element affects template boundary element function, telomere sequence and shelterin binding. Proc. Natl. Acad. Sci. USA 2015, 112, 11312–11317. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.J.; Theimer, C.A.; Finger, L.D.; Feigon, J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006, 34, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Cash, D.D.; Feigon, J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 2017, 45, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Collins, K. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. USA 2002, 99, 6865–6890. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.J.; Akiyama, B.M.; Stone, M.D.; Cech, T.R. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat. Struct. Mol. Biol. 2011, 18, 1371–1375. [Google Scholar] [CrossRef] [PubMed]
- McCormick-Graham, M.; Romero, D.P. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol. Cell. Biol. 1996, 16, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Egan, E.D.; Collins, K. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol. Cell. Biol. 2012, 32, 2428–2439. [Google Scholar] [CrossRef] [PubMed]
- Stellwagen, A.E.; Haimberger, Z.W.; Veatch, J.R.; Gottschling, D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003, 17, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Witkin, K.L.; Collins, K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004, 18, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Collins, K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell. Biol. 2006, 26, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.J.; Gooding, A.R.; Cech, T.R. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol. Cell. Biol. 2010, 30, 4965–4976. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Wang, Z.; Koo, B.K.; Patel, A.; Cascio, D.; Collins, K.; Feigon, J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell 2012, 47, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Chapon, C.; Cech, T.R.; Zaug, A.J. Polyadenylation of telomerase RNA in budding yeast. RNA 1997, 3, 1337–1351. [Google Scholar] [PubMed]
- Leonardi, J.; Box, J.A.; Bunch, J.T.; Baumann, P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008, 15, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Bosoy, D.; Peng, Y.; Mian, I.S.; Lue, N.F. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003, 278, 3882–3890. [Google Scholar] [CrossRef] [PubMed]
- Kuehner, J.N.; Pearson, E.L.; Moore, C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell. Biol. 2011, 12, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.F.; Larose, S.; Abou Elela, S.; Wellinger, R.J. Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res. 2012, 40, 5625–5636. [Google Scholar] [CrossRef] [PubMed]
- Jamonnak, N.; Creamer, T.J.; Darby, M.M.; Schaughency, P.; Wheelan, S.J.; Corden, J.L. Yeast Nrd1, Nab3 and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 2011, 17, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Box, J.A.; Bunch, J.T.; Tang, W.; Baumann, P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 2008, 456, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Zaug, A.J.; Sobel, S.G.; Wolin, S.L.; Cech, T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 1999, 401, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.T.; Forstemann, K.; Gasser, S.M.; Lingner, J. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002, 3, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, F.; Olivier, C.; Dandjinou, A.T.; Wellinger, R.J.; Chartrand, P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008, 27, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Kannan, R.; Blanchette, M.; Baumann, P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 2012, 484, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Sexton, A.N.; Collins, K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011, 31, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Meier, U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004, 23, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Kiss, A.M.; Darzacq, X.; Kiss, T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 2006, 26, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Darzacq, X.; Kittur, N.; Roy, S.; Shav-Tal, Y.; Singer, R.H.; Meier, U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006, 173, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Marmier-Gourrier, N.; Pradet-Balade, B.; Wurth, L.; Verheggen, C.; Jady, B.E.; Rothe, B.; Pescia, C.; Robert, M.C.; Kiss, T.; et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008, 180, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Isaac, C.; Wang, C.; Dragon, F.; Pogacic, V.; Meier, U.T. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Cell. Biol. 2000, 11, 567–577. [Google Scholar] [CrossRef]
- Jady, B.E.; Bertrand, E.; Kiss, T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell. Biol. 2004, 164, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Collins, K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006, 20, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Savage, S.A.; Shkreli, M.; Giri, N.; Jessop, L.; Myers, T.; Chen, R.; Alter, B.P.; Artandi, S.E. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenital. Genes Dev. 2011, 25, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Tesmer, V.M.; Savre-Train, I.; Shay, J.W.; Wright, W.E. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999, 19, 3989–3997. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, R.L.; Ziegler, T.D.; Supakorndej, T.; Terns, R.M.; Terns, M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 2006, 17, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Collins, K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008, 129, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.E.; Aisner, D.L.; Baur, J.; Tesmer, V.M.; Dy, M.; Ouellette, M.; Trager, J.B.; Morin, G.B.; Toft, D.O.; Shay, J.W.; et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999, 13, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, H.L.; Jarvis, J.L.; Turner, J.W.; Elmore, L.W.; Holt, S.E. Stable association of hsp90 and p23 but Not hsp70, with active human telomerase. J. Biol. Chem. 2001, 276, 15571–15574. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Park, S.M.; Kang, M.R.; Oh, S.Y.; Lee, T.H.; Muller, M.T.; Chung, I.K. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005, 19, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.E.; Yu, E.Y.; Cho, C.H.; Lee, J.; Muller, M.T.; Chung, I.K. DNA-protein kinase catalytic subunit-interacting protein KIP binds telomerase by interacting with human telomerase reverse transcriptase. J. Biol. Chem. 2004, 279, 34750–34755. [Google Scholar] [CrossRef] [PubMed]
- Bachand, F.; Boisvert, F.M.; Cote, J.; Richard, S.; Autexier, C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol. Biol. Cell. 2002, 13, 3192–3202. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.E.; Kugrens, P. Relationship between the flagellates and the ciliates. Microbiol. Rev. 1992, 56, 529–542. [Google Scholar] [PubMed]
- Cano, M.I.; Dungan, J.M.; Agabian, N.; Blackburn, E.H. Telomerase in kinetoplastid parasitic protozoa. Proc. Natl. Acad. Sci. USA 1999, 96, 3616–3621. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, E.J.; Nunes, V.S.; da Silva, M.S.; Segatto, M.; Myler, P.J.; Cano, M.I. The putative Leishmania telomerase RNA (LeishTER) undergoes trans-splicing and contains a conserved template sequence. PLoS ONE 2014, 9, e112061. [Google Scholar] [CrossRef] [PubMed]
- Avilion, A.A.; Harrington, L.A.; Greider, C.W. Tetrahymena telomerase RNA levels increase during macronuclear development. Dev. Genet. 1992, 13, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Cech, T.R. Telomerase RNA localized in the replication band and spherical subnuclear organelles in hypotrichous ciliates. J. Cell Biol. 1995, 130, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Collins, K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 2009, 36, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Collins, K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 2010, 285, 16434–16443. [Google Scholar] [CrossRef] [PubMed]
- Linger, B.R.; Morin, G.B.; Price, C.M. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol. Biol. Cell 2011, 22, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.A.; Wuttke, D.S. Telomerase and telomere-associated proteins: Structural insights into mechanism and evolution. Structure 2012, 20, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, N.R.; Dickey, T.H.; Hom, R.A.; Wuttke, D.S. Tying up the Ends: Plasticity in the Recognition of Single-Stranded DNA at Telomeres. Biochemistry 2016, 55, 5326–5340. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Cervantes, R.B.; Mandell, E.K.; Otero, J.H.; Lundblad, V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007, 14, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.K.; Lundblad, V. Est1 and Cdc13 as comediators of telomerase access. Science 1999, 286, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.S.; Taggart, A.K.; Zakian, V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004, 11, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Pfingsten, J.S.; Goodrich, K.J.; Taabazuing, C.; Ouenzar, F.; Chartrand, P.; Cech, T.R. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 2012, 148, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.A.; Nakamura, T.M. Protection and replication of telomeres in fission yeast. Biochem. Cell Biol. 2009, 87, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, J.; Jun, H.I.; Kim, J.K.; Qiao, F. Multi-step coordination of telomerase recruitment in fission yeast through two coupled telomere-telomerase interfaces. Elife 2016, 5, e15470. [Google Scholar] [CrossRef] [PubMed]
- Abreu, E.; Aritonovska, E.; Reichenbach, P.; Cristofari, G.; Culp, B.; Terns, R.M.; Lingner, J.; Terns, M.P. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010, 30, 2971–2982. [Google Scholar] [CrossRef] [PubMed]
- Tejera, A.M.; Stagno d’Alcontres, M.; Thanasoula, M.; Marion, R.M.; Martinez, P.; Liao, C.; Flores, J.M.; Tarsounas, M.; Blasco, M.A. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming and normal skin development in mice. Dev. Cell 2010, 18, 775–789. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Chaiken, M.F.; Wang, F.; Price, C.M. Maintaining the end: Roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 2012, 730, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Wan, M.; Safari, A.; Kim, H.; Sun, W.; O’Connor, M.S.; Songyang, Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 2007, 445, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Ford, L.P.; Lenertz, L.; Wright, W.E.; Shay, J.W. Human Ku70/80 associates physically with telomerase through interaction with Htert. J. Biol. Chem. 2002, 277, 47242–47247. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Jung, D.; Jung, Y.; Lee, S.G.; Lee, I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000, 481, 81–85. [Google Scholar] [CrossRef]
- Cano, M.I.; Blake, J.J.; Blackburn, E.H.; Agabian, N. A Trypanosoma brucei protein complex that binds G-overhangs and co-purifies with telomerase activity. J. Biol. Chem. 2002, 277, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Zou, Y.; Hiyama, E.; Wright, W.E. Telomerase and cancer. Hum. Mol. Genet. 2001, 10, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Nelson, A.D.; Shippen, D.E. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol. Cell. Biol. 2008, 28, 2332–2341. [Google Scholar] [CrossRef] [PubMed]
- Livengood, A.J.; Zaug, A.J.; Cech, T.R. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol. Cell Biol. 2002, 22, 2366–2374. [Google Scholar] [CrossRef]
- Cifuentes-Rojas, C.; Shippen, D.E. Telomerase regulation. Mutat. Res. 2012, 730, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Podell, E.R.; Nandakumar, J.; Cech, T.R. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010, 24, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Collins, K. Control of telomerase action at human telomeres. Nat. Struct. Mol. Biol. 2015, 22, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gu, P.; Wu, J.; Chen, X.; Niu, S.; Sun, H.; Wu, L.; Li, N.; Peng, J.; Shi, S.; et al. Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat. Commun. 2017, 8, 14929. [Google Scholar] [CrossRef] [PubMed]
- Marcand, S.; Brevet, V.; Mann, C.; Gilson, E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000, 10, 487–490. [Google Scholar] [CrossRef]
- Diede, S.J.; Gottschling, D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 1999, 99, 723–733. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Vaziri, H.; Patterson, C.; Goldstein, S.; Younglai, E.V.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 1992, 89, 10114–10118. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Stansel, R.M.; de Lange, T.; Griffith, J.D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001, 20, 5532–5540. [Google Scholar] [CrossRef] [PubMed]
- Herbig, U.; Jobling, W.A.; Chen, B.P.; Chen, D.J.; Sedivy, J.M. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53 and p21(CIP1) but not p16(INK4a). Mol. Cell 2004, 14, 501–513. [Google Scholar] [CrossRef]
- D’ADDA di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Pereira-Smith, O.M.; Wright, W.E. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991, 196, 33–39. [Google Scholar] [CrossRef]
- Artandi, S.E.; DePinho, R.A. Telomeres and telomerase in cancer. Carcinogenesis 2010, 31, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Shay, J.W. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 1992, 27, 383–389. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008, 129, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Johnson, J.Z.; Vogan, J.M.; Wagner, T.; Boyle, J.M.; Hockemeyer, D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife 2015, 4, e07918. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.P.; Lee, L.W.; Tang, S.C.; Hsin, I.L.; Lin, Y.W.; Ko, J.L. Epidermal growth factor activates telomerase activity by direct binding of Ets-2 to hTERT promoter in lung cancer cells. Tumour Biol. 2015, 36, 5389–5398. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Rachakonda, P.S.; Hemminki, K.; Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014, 24, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Ccancer 2015, 51, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Baena-Del Valle, J.A.; Zheng, Q.; Esopi, D.M.; Rubenstein, M.; Hubbard, G.K.; Moncaliano, M.C.; Hruszkewycz, A.; Vaghasia, A.; Yegnasubramanian, S.; Wheelan, S.J.; et al. MYC drives overexpression of telomerase RNA (hTR/TERC) in prostate cancer. J. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Akincilar, S.C.; Unal, B.; Tergaonkar, V. Reactivation of telomerase in cancer. Cell Mol. Life Sci. 2016, 73, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Dokal, I. Dyskeratosis congenita: Advances in the understanding of the telomerase defect and the role of stem cell transplantation. Pediatr. Transplant. 2007, 11, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.R.; Wood, E.; Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 1999, 402, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Savage, S.A.; Giri, N.; Baerlocher, G.M.; Orr, N.; Lansdorp, P.M.; Alter, B.P. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008, 82, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Walne, A.J.; Vulliamy, T.; Marrone, A.; Beswick, R.; Kirwan, M.; Masunari, Y.; Al-Qurashi, F.H.; Aljurf, M.; Dokal, I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007, 16, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Vulliamy, T.; Beswick, R.; Kirwan, M.; Marrone, A.; Digweed, M.; Walne, A.; Dokal, I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2008, 105, 8073–8078. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; Collins, K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006, 20, 2848–2858. [Google Scholar] [CrossRef] [PubMed]
- Rocha, V.; Devergie, A.; Socie, G.; Ribaud, P.; Esperou, H.; Parquet, N.; Gluckman, E. Unusual complications after bone marrow transplantation for dyskeratosis congenita. Br. J. Harmatol. 1998, 103, 243–248. [Google Scholar] [CrossRef]
- Brouilette, S.W.; Moore, J.S.; McMahon, A.D.; Thompson, J.R.; Ford, I.; Shepherd, J.; Packard, C.J.; Samani, N.J. Telomere length, risk of coronary heart disease and statin treatment in the West of Scotland Primary Prevention Study: A nested case-control study. Lancet 2007, 369, 107–114. [Google Scholar] [CrossRef]
- Armanios, M. Syndromes of telomere shortening. Annu. Rev. Genom. Hum. Genet. 2009, 10, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Hemann, M.T.; Rudolph, K.L.; Strong, M.A.; DePinho, R.A.; Chin, L.; Greider, C.W. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell 2001, 12, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Cross, G.A. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 1975, 71, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Van der Ploeg, L.H.; Valerio, D.; De Lange, T.; Bernards, A.; Borst, P.; Grosveld, F.G. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 1982, 10, 5905–5923. [Google Scholar] [CrossRef] [PubMed]
- Berriman, M.; Ghedin, E.; Hertz-Fowler, C.; Blandin, G.; Renauld, H.; Bartholomeu, D.C.; Lennard, N.J.; Caler, E.; Hamlin, N.E.; Haas, B.; et al. The genome of the African trypanosome Trypanosoma brucei. Science 2005, 309, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Pays, E.; Guyaux, M.; Aerts, D.; Van Meirvenne, N.; Steinert, M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature 1985, 316, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Myler, P.; Nelson, R.G.; Agabian, N.; Stuart, K. Two mechanisms of expression of a predominant variant antigen gene of Trypanosoma brucei. Nature 1984, 309, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Dreesen, O.; Li, B.; Cross, G.A. Telomere structure and function in trypanosomes: A proposal. Nat. Rev. Microbiol. 2007, 5, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Dreesen, O.; Cross, G.A. Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol. Cell. Biol. 2006, 26, 4911–4919. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Figueiredo, L.M.; Espinal, A.; Okubo, E.; Li, B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 2009, 137, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.M.; Pirrit, L.A.; Scherf, A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000, 106, 169–174. [Google Scholar] [CrossRef]
- Scherf, A.; Figueiredo, L.M.; Freitas-Junior, L.H. Plasmodium telomeres: A pathogen’s perspective. Curr. Opin. Microbiol. 2001, 4, 409–414. [Google Scholar] [CrossRef]
- Smith, J.D.; Chitnis, C.E.; Craig, A.G.; Roberts, D.J.; Hudson-Taylor, D.E.; Peterson, D.S.; Pinches, R.; Newbold, C.I.; Miller, L.H. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 1995, 82, 101–110. [Google Scholar] [CrossRef]
- Freitas-Junior, L.H.; Hernandez-Rivas, R.; Ralph, S.A.; Montiel-Condado, D.; Ruvalcaba-Salazar, O.K.; Rojas-Meza, A.P.; Mancio-Silva, L.; Leal-Silvestre, R.J.; Gontijo, A.M.; Shorte, S.; et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 2005, 121, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Duraisingh, M.T.; Voss, T.S.; Marty, A.J.; Duffy, M.F.; Good, R.T.; Thompson, J.K.; Freitas-Junior, L.H.; Scherf, A.; Crabb, B.S.; Cowman, A.F. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 2005, 121, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, M.; Sobel, R.E.; Allis, C.D.; Turner, B.M.; Broach, J.R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 1996, 16, 4349–4356. [Google Scholar] [CrossRef] [PubMed]
- Shankaranarayana, G.D.; Motamedi, M.R.; Moazed, D.; Grewal, S.I. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 2003, 13, 1240–1246. [Google Scholar] [CrossRef]
- Religa, A.A.; Ramesar, J.; Janse, C.J.; Scherf, A.; Waters, A.P. P. berghei telomerase subunit TERT is essential for parasite survival. PLoS ONE 2014, 9, e108930. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.; Dey, A.; Xu, L.; Sanford, S.; Tain, L.; Weizmann, Y.; Chakrabarti, K. Role of telomerase RNP in telomere chromatin remodeling in Plasmodium falciparum. 2018; manuscript in preparation. [Google Scholar]
- Gehring, W.J.; Klemenz, R.; Weber, U.; Kloter, U. Functional analysis of the white gene of Drosophila by P-factor-mediated transformation. EMBO J. 1984, 3, 2077–2085. [Google Scholar] [PubMed]
- Hazelrigg, T.; Levis, R.; Rubin, G.M. Transformation of white locus DNA in drosophila: Dosage compensation, zeste interaction and position effects. Cell 1984, 36, 469–481. [Google Scholar] [CrossRef]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef]
- Aparicio, O.M.; Billington, B.L.; Gottschling, D.E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 1991, 66, 1279–1287. [Google Scholar] [CrossRef]
- Renauld, H.; Aparicio, O.M.; Zierath, P.D.; Billington, B.L.; Chhablani, S.K.; Gottschling, D.E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength and by SIR3 dosage. Genes Dev. 1993, 7, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Kyrion, G.; Liu, K.; Liu, C.; Lustig, A.J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993, 7, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Eugster, A.; Lanzuolo, C.; Bonneton, M.; Luciano, P.; Pollice, A.; Pulitzer, J.F.; Stegberg, E.; Berthiau, A.S.; Forstemann, K.; Corda, Y.; et al. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat. Struct. Mol. Biol. 2006, 13, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Zou, Y.; Shay, J.W.; Wright, W.E. Telomere position effect in human cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef] [PubMed]
- Koering, C.E.; Pollice, A.; Zibella, M.P.; Bauwens, S.; Puisieux, A.; Brunori, M.; Brun, C.; Martins, L.; Sabatier, L.; Pulitzer, J.F.; et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002, 3, 1055–1061. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, A.; Chakrabarti, K. Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. Int. J. Mol. Sci. 2018, 19, 333. https://doi.org/10.3390/ijms19020333
Dey A, Chakrabarti K. Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. International Journal of Molecular Sciences. 2018; 19(2):333. https://doi.org/10.3390/ijms19020333
Chicago/Turabian StyleDey, Abhishek, and Kausik Chakrabarti. 2018. "Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites" International Journal of Molecular Sciences 19, no. 2: 333. https://doi.org/10.3390/ijms19020333
APA StyleDey, A., & Chakrabarti, K. (2018). Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. International Journal of Molecular Sciences, 19(2), 333. https://doi.org/10.3390/ijms19020333





