Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice
Abstract
1. Introduction
2. Results
2.1. Identification of Porcine Myh1 Gene
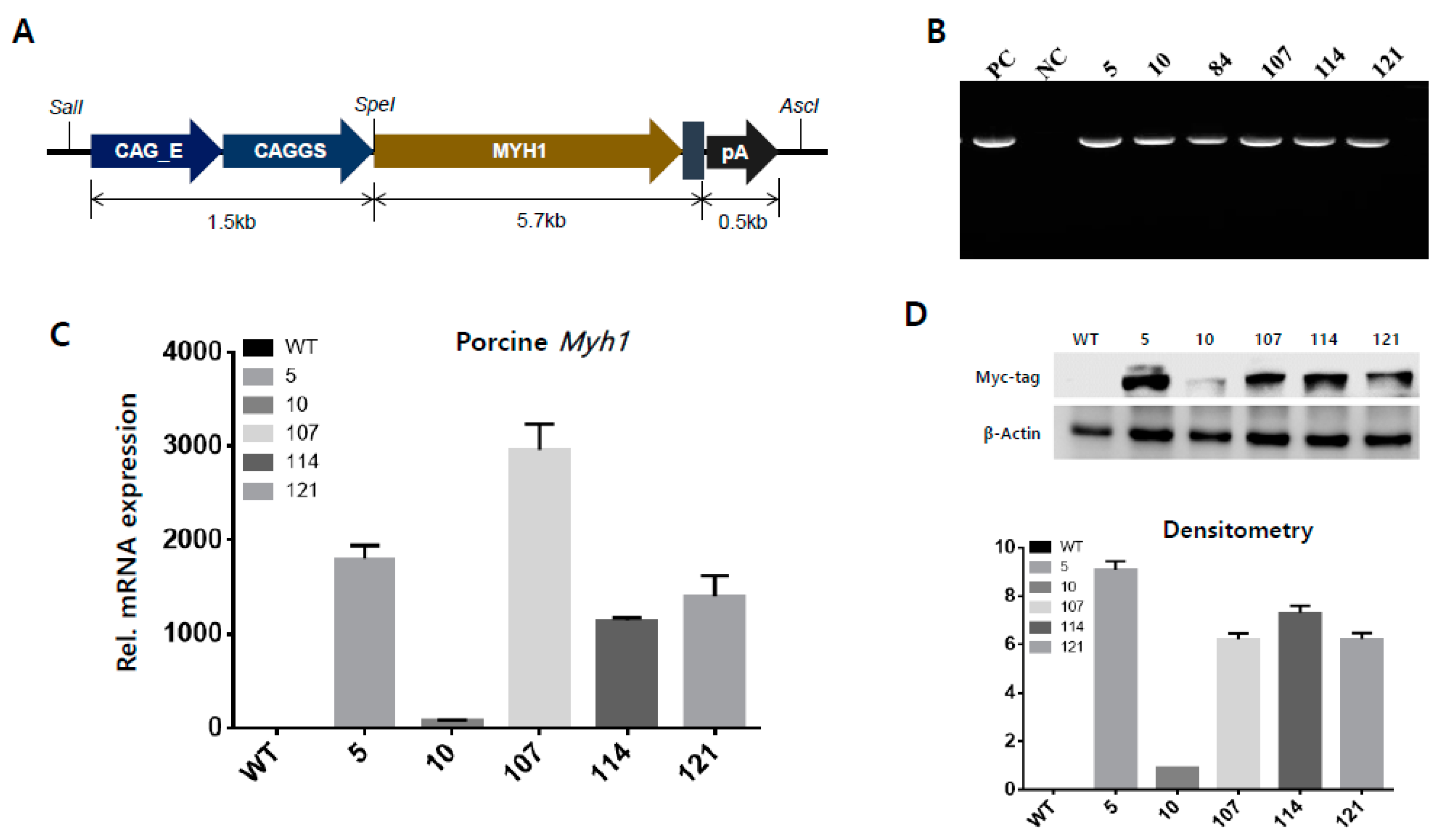
2.2. Establishment of Ectopic Overexpression of Porcine Myh1 in Mice
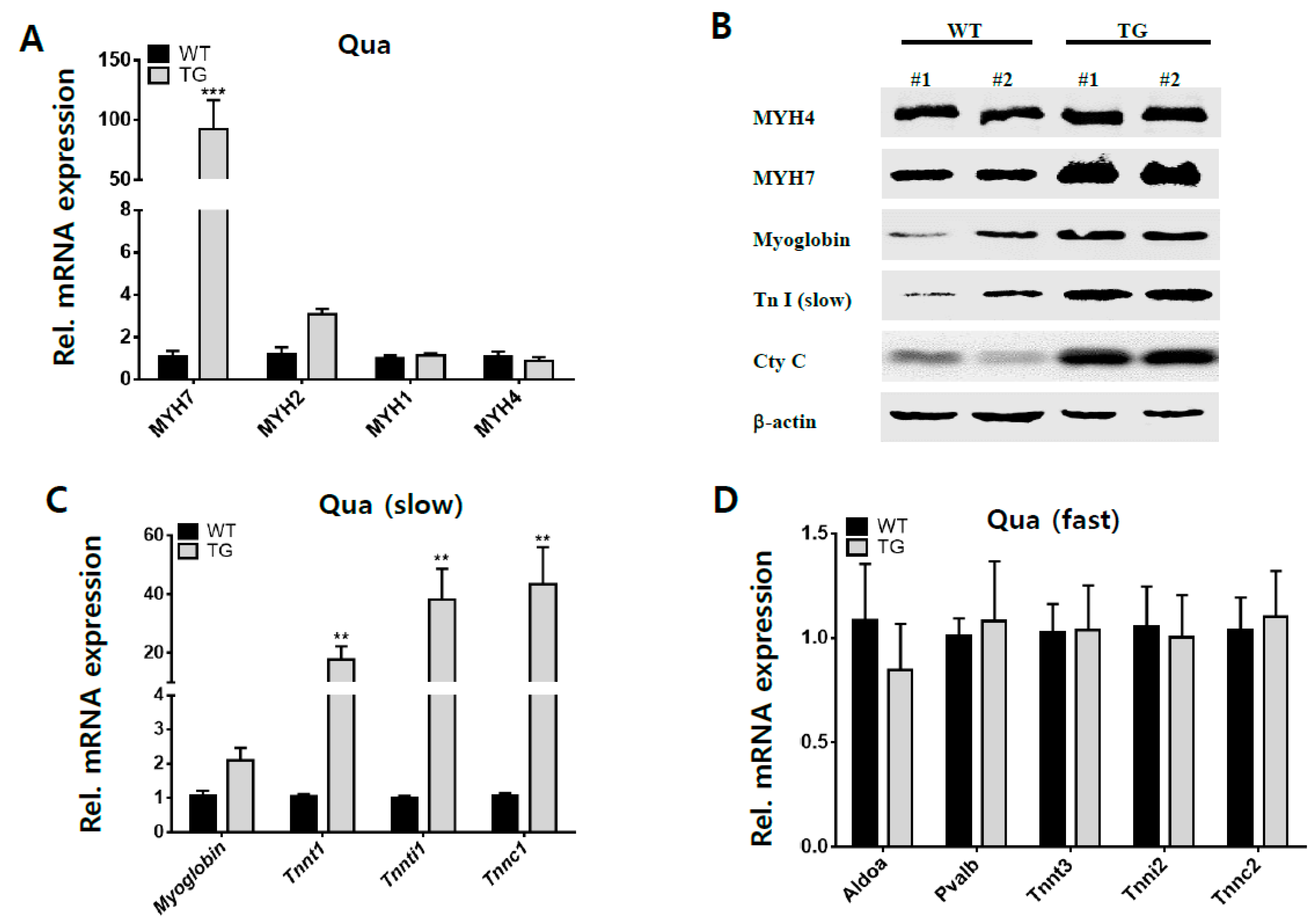
2.3. Effects on Slow Fiber Muscle in Myh1 TG Mice
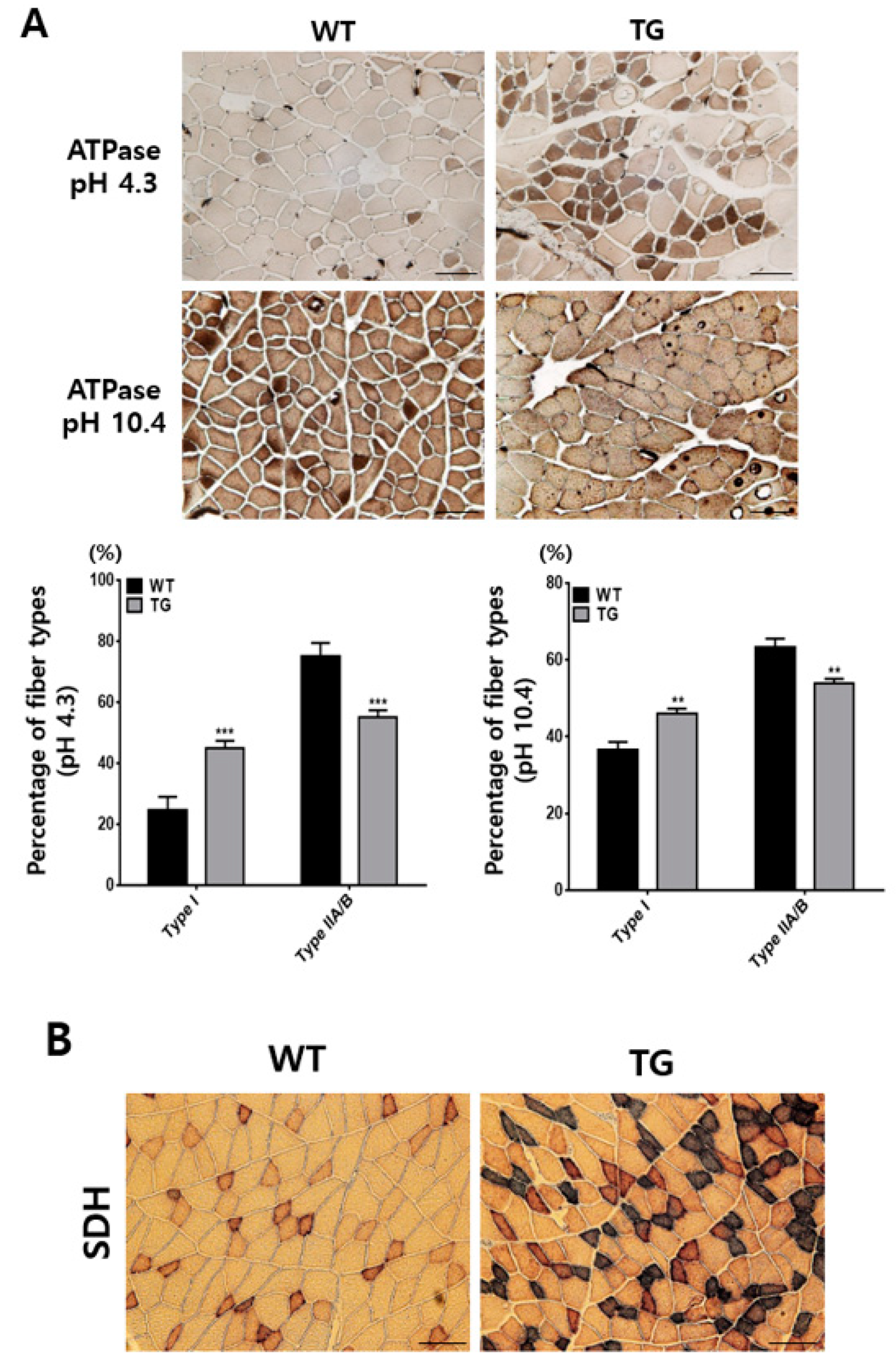
2.4. Effect of Ectopic Expressed Porcine Myh1 to Muscle Type Formation
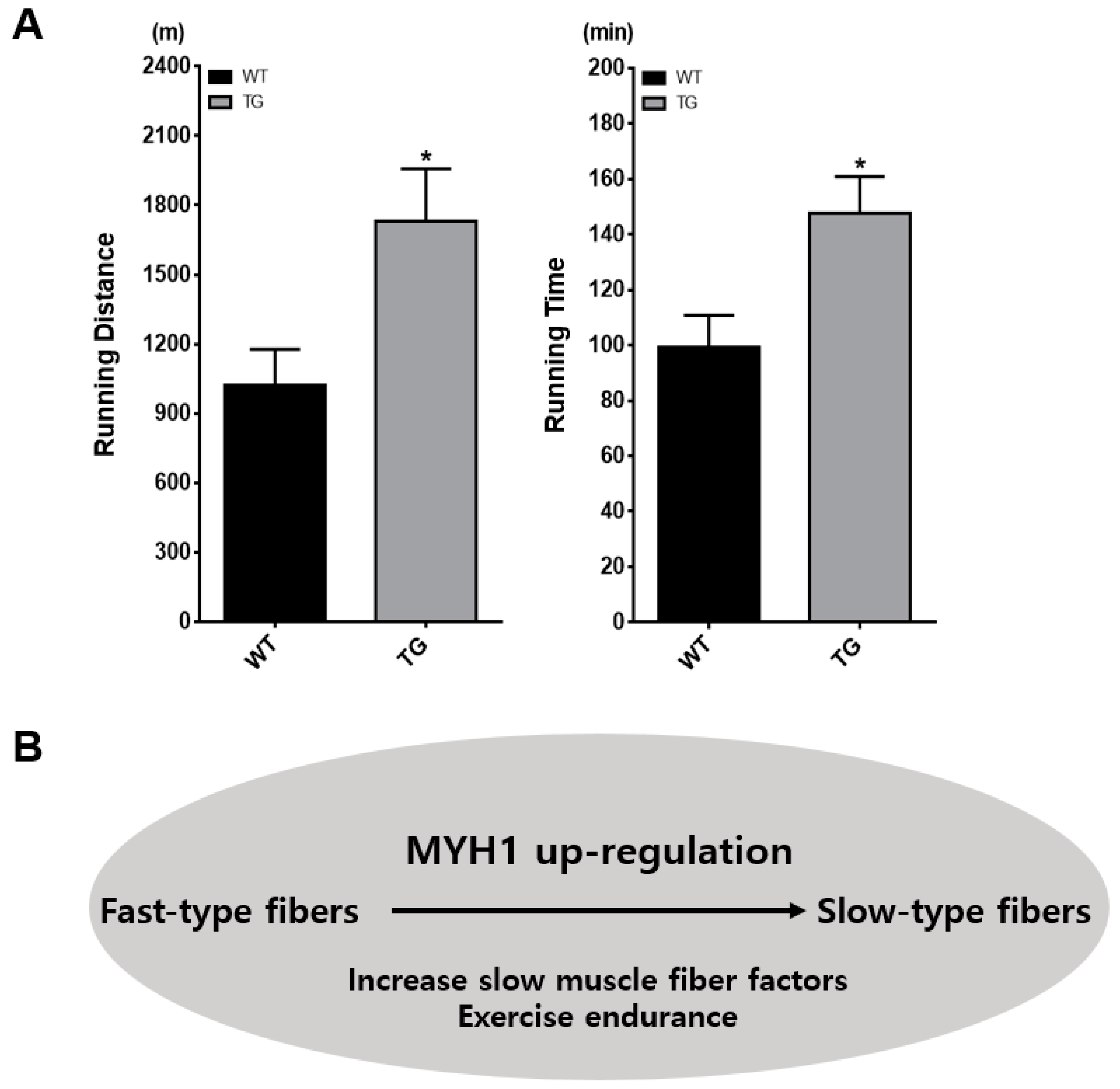
2.5. Enhanced Physical Performance in Myh1 TG Mice
3. Discussion
4. Materials and Methods
4.1. Use of Mice and IACUC Commitment
4.2. Construct of Myh1 Overexpression Vector and TG Mice Production
4.3. Analysis of Gene Expression
4.4. Western Blotting
4.5. Immunohistochemistry
4.6. ATPase and Succinate Dehydrogenase Staining
4.7. Physiological Studies
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pette, D. The adaptive potential of skeletal muscle fibers. Can. J. Appl. Physiol. 2002, 27, 423–448. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 1996, 76, 371–423. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed]
- Booth, F.W.; Thomason, D.B. Molecular and cellular adaptation of muscle in response to exercise: Perspectives of various models. Physiol. Rev. 1991, 71, 541–585. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, N. Muscle fibre types: Their role in health, disease and as therapeutic targets. OA Biol. 2013, 1, 1–7. [Google Scholar]
- Berchtold, M.W.; Brinkmeier, H.; Müntener, M. Calcium Ion in Skeletal Muscle: Its Crucial Role for Muscle Function, Plasticity, and Disease. Physiol. Rev. 2000, 80, 1215–1265. [Google Scholar] [CrossRef] [PubMed]
- Greising, S.M.; Gransee, H.M.; Mantilla, C.B.; Sieck, G.C. Systems biology of skeletal muscle: Fiber type as an organizing principle. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling Pathways in Skeletal Muscle Remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Augusto, V.; Padovani, C.R.; Eduardo, G.; Campos, R. Skeletal Muscle Fiber Types in C57Bl6J Mice. Braz. J. Morphol. Sci. 2004, 21, 89–94. [Google Scholar]
- Arany, Z.; Lebrasseur, N.; Morris, C.; Smith, E.; Yang, W.; Ma, Y.; Chin, S.; Spiegelman, B.M. The Transcriptional Coactivator PGC-1β Drives the Formation of Oxidative Type IIX Fibers in Skeletal Muscle. Cell Metab. 2007, 5, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, H.; Tarr, P.T.; Zhang, C.Y.; Wu, Z.; Boss, O.; Michael, L.F.; Puigserver, P.; Isotani, E.; Olson, E.N.; et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 2002, 418, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Hering, T.; Braubach, P.; Landwehrmeyer, G.B.; Lindenberg, K.S.; Melzer, W. Fast-to-slow transition of skeletal muscle contractile function and corresponding changes in myosin heavy and light chain formation in the R6/2 mouse model of Huntington’s Disease. PLoS ONE 2016, 11, e0166106. [Google Scholar] [CrossRef] [PubMed]
- Mithal, A.; Bonjour, J.P.; Boonen, S.; Burckhardt, P.; Degens, H.; El Hajj Fuleihan, G.; Josse, R.; Lips, P.; Morales Torres, J.; Rizzoli, R.; et al. Impact of nutrition on muscle mass, strength, and performance in older adults. Osteoporos. Int. 2013, 24, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Edström, L.; Lindegren, B.; Gorza, L.; Schiaffino, S. MHC composition and enzyme-histochemical and physiological properties of a novel fast-twitch motor unit type. Am. J. Physiol. 1991, 261, C93–C101. [Google Scholar] [CrossRef] [PubMed]
- Jacobs-El, J.; Ashley, W.; Russell, B. IIx and slow myosin expression follow mitochondrial increases in transforming muscle fibers. Am. J. Physiol. 1993, 265, C79–C84. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liu, X.; Zhou, L.; Yang, J.; Yang, B.; Ma, H.; Xie, X.; Huang, Y.; Fang, S.; Xiao, S.; et al. Genome-wide association analysis reveals genetic loci and candidate genes for meat quality traits in Chinese Laiwu pigs. Mamm. Genome 2015, 26, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How muscle structure and composition influence meat and flesh quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.; Hardeman, E. Multiple mechanisms regulate muscle fiber diversity. FASEB J. 1991, 5, 3064–3070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Zhang, C.L.; Yu, R.T.; Cho, H.K.; Nelson, M.C.; Bayuga-Ocampo, C.R.; Ham, J.; Kang, H.; Evans, R.M. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2004, 2, e294. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Meek, T.H.; Pandey, S.N.; Broitman-Maduro, G.; Maduro, M.F.; Bronikowski, A.M.; Garland, T.; Chen, Y.-W. Gene expression profiling of gastrocnemius of “minimuscle” mice. Physiol. Genom. 2013, 45, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Leinwand, L.A. the Mammalian Myosin Heavy Chain Gene Family. Annu. Rev. Cell Dev. Biol. 1996, 12, 417–439. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Carlsen, H.; Gundersen, K. Helix-loop-helix transcription factors in electrically active and inactive skeletal muscles. Muscle Nerve 2000, 23, 1374–1380. [Google Scholar] [CrossRef]
- Luo, W.; Cheng, D.; Chen, S.; Wang, L.; Li, Y.; Ma, X.; Song, X.; Liu, X.; Li, W.; Liang, J.; et al. Genome-wide association analysis of meat quality traits in a porcine Large White × Minzhu intercross population. Int. J. Biol. Sci. 2012, 8, 580–595. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.; Maves, L. Skeletal muscle fibre type: Using insights from muscle developmetal biology to dissect targets for susceptibility ans resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 2017, 5, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequence (5’-3’) | Size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| Myh7 (mouse) | AGTCCCAGGTCAACAAGCTG | TTCCACCTAAAGGGCTGTTG | 145 |
| Myh2 (mouse) | AGTCCCAGGTCAACAAGCTG | GCATGACCAAAGGTTTCACA | 130 |
| Myh1 (mouse) | AGTCCCAGGTCAACAAGCTG | CACATTTTGCTCATCTCTTTG | 113 |
| Myh1 (porcine) | CTTCACTGGCGCAGCAGGT | AGATGCGGATGCCCTCCA | 256 |
| Myh4 (mouse) | AGTCCCAGGTCAACAAGCTG | TTTCTCCTGTCACCTCTCAACA | 100 |
| Myoglobin (mouse) | GCAAGGCCCTGGAGCTCTTC | GCTTGGTGGGCTGGACAGTG | 100 |
| Tnnt1 (mouse) | CCCCCGAAGATTCCAGAAGG | TGCGGTCTTTTAGTGCAATGAG | 154 |
| Tnni1 (mouse) | ATGCCGGAAGTTGAGAGGAAA | TCCGAGAGGTAACGCACCTT | 140 |
| Tnnc1 (mouse) | GCGGTAGAACAGTTGACAGAG | CCAGCTCCTTGGTGCTGAT | 103 |
| Aldoa (mouse) | GGCAACACCCAGCAACAGAC | TCATCTGCAGCCAGGATGCC | 158 |
| Pvalb (mouse) | ATCAAGAAGGCGATAGGAGCC | GGCCAGAAGCGTCTTTGTT | 252 |
| Tnnt3 (mouse) | GGAACGCCAGAACAGATTGG | TGGAGGACAGAGCCTTTTTCTT | 104 |
| Tnni2 (mouse) | AGAGTGTGATGCTCCAGATAGC | AGCAACGTCGATCTTCGCA | 170 |
| Tnnc2 (mouse) | ATGGCAGCGGTACTATCGACT | CCTTCGCATCCTCTTTCATCTG | 72 |
| GAPDH (mouse) | TGAAGGTCGGTGTGAACGG | CGTGAGTGGAGTCATACTGGAA | 150 |
| GAPDH (porcine) | CCTTCATTGACCTCAACTACAT | CCAAAGTTGTCATGGATGACC | 400 |
| Antibody | Number | Source |
|---|---|---|
| Myc-Tag | #2276 | Cell Signaling |
| MYH4 | SC-168672 | Santa Cruz Biotechnology |
| MYH7 | SC-53089 | Santa Cruz Biotechnology |
| Myoglobin | SC-8081 | Santa Cruz Biotechnology |
| Troponin I-SS | SC-8119 | Santa Cruz Biotechnology |
| Cytochrom C | SC-13156 | Santa Cruz Biotechnology |
| β-actin | #4970 | Cell Signaling |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, J.S.; Kim, D.-H.; Park, H.-B.; Han, S.-H.; Hwang, S.; Cho, I.-C.; Lee, J.-W. Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice. Int. J. Mol. Sci. 2018, 19, 2959. https://doi.org/10.3390/ijms19102959
Ahn JS, Kim D-H, Park H-B, Han S-H, Hwang S, Cho I-C, Lee J-W. Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice. International Journal of Molecular Sciences. 2018; 19(10):2959. https://doi.org/10.3390/ijms19102959
Chicago/Turabian StyleAhn, Jin Seop, Dong-Hwan Kim, Hee-Bok Park, Sang-Hyun Han, Seongsoo Hwang, In-Cheol Cho, and Jeong-Woong Lee. 2018. "Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice" International Journal of Molecular Sciences 19, no. 10: 2959. https://doi.org/10.3390/ijms19102959
APA StyleAhn, J. S., Kim, D.-H., Park, H.-B., Han, S.-H., Hwang, S., Cho, I.-C., & Lee, J.-W. (2018). Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice. International Journal of Molecular Sciences, 19(10), 2959. https://doi.org/10.3390/ijms19102959






