Proteomic Analysis and Identification of Possible Allergenic Proteins in Mature Pollen of Populus tomentosa
Abstract
:1. Introduction
2. Results and Discussion
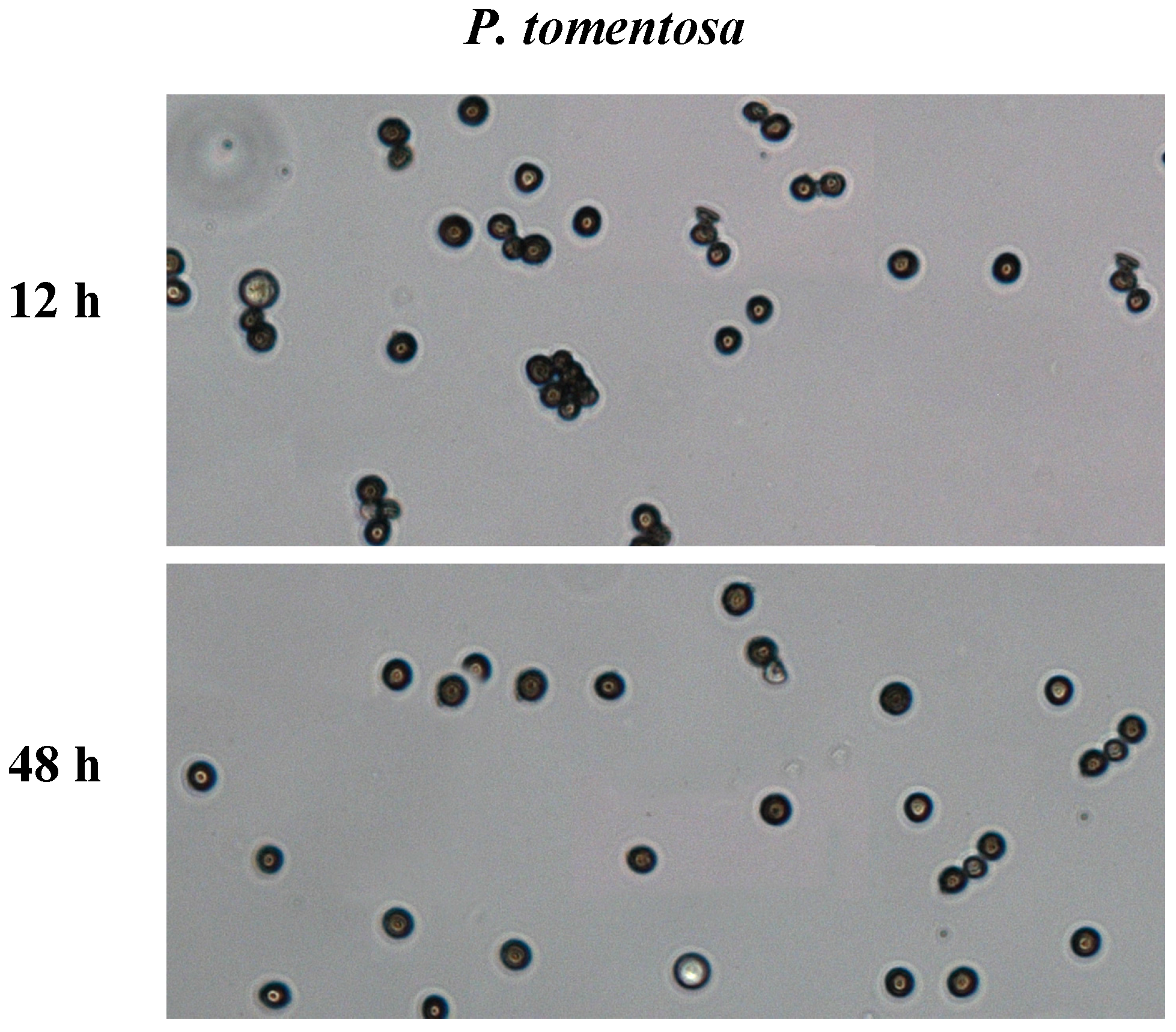
2.1. In Vitro Pollen Germination
2.2. Proteomic Maps of P. tomentosa Mature Pollen
2.3. Functional Classification of Identified Proteins
2.4. Prediction of Allergens in P. tomentosa Mature Pollen
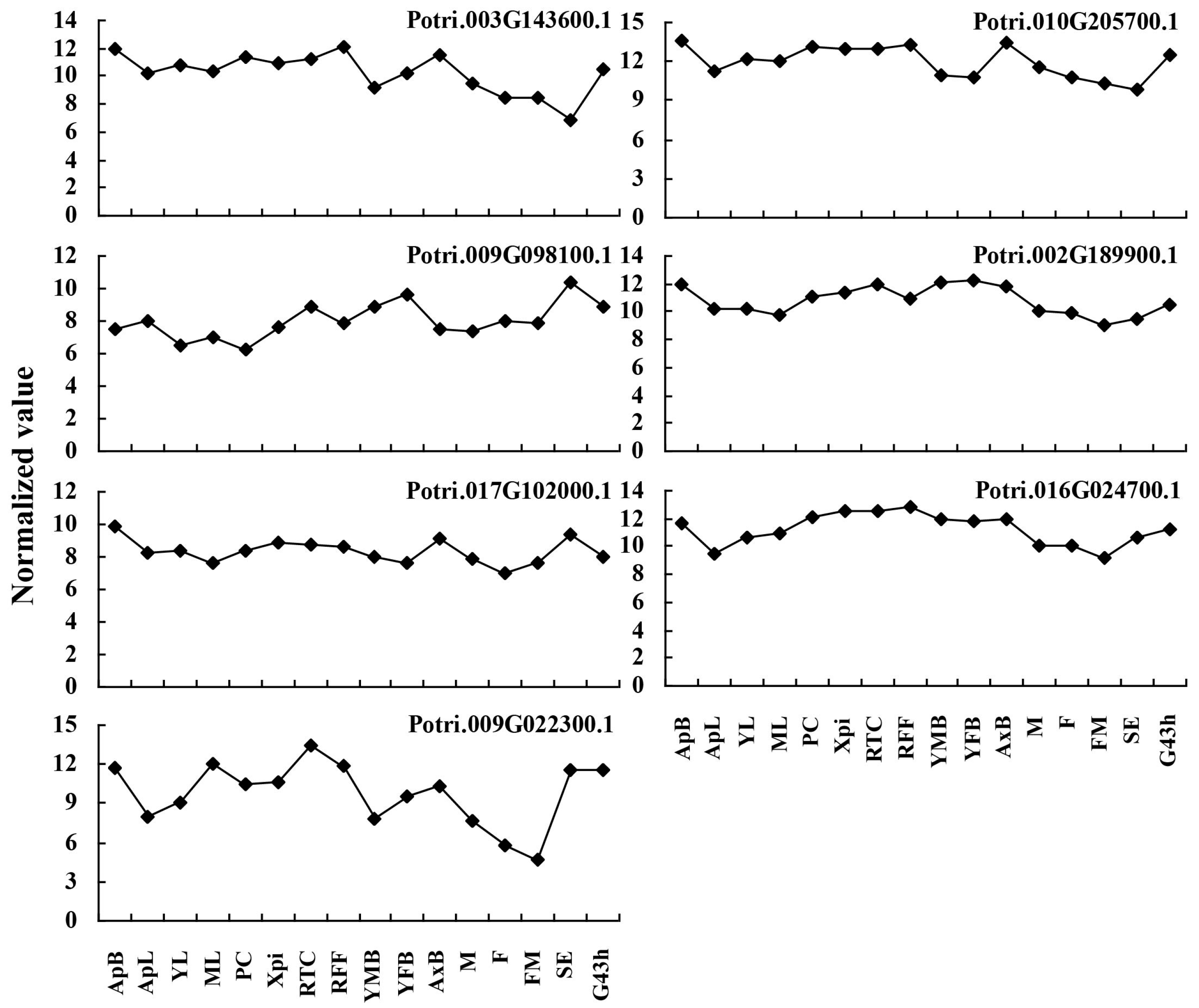
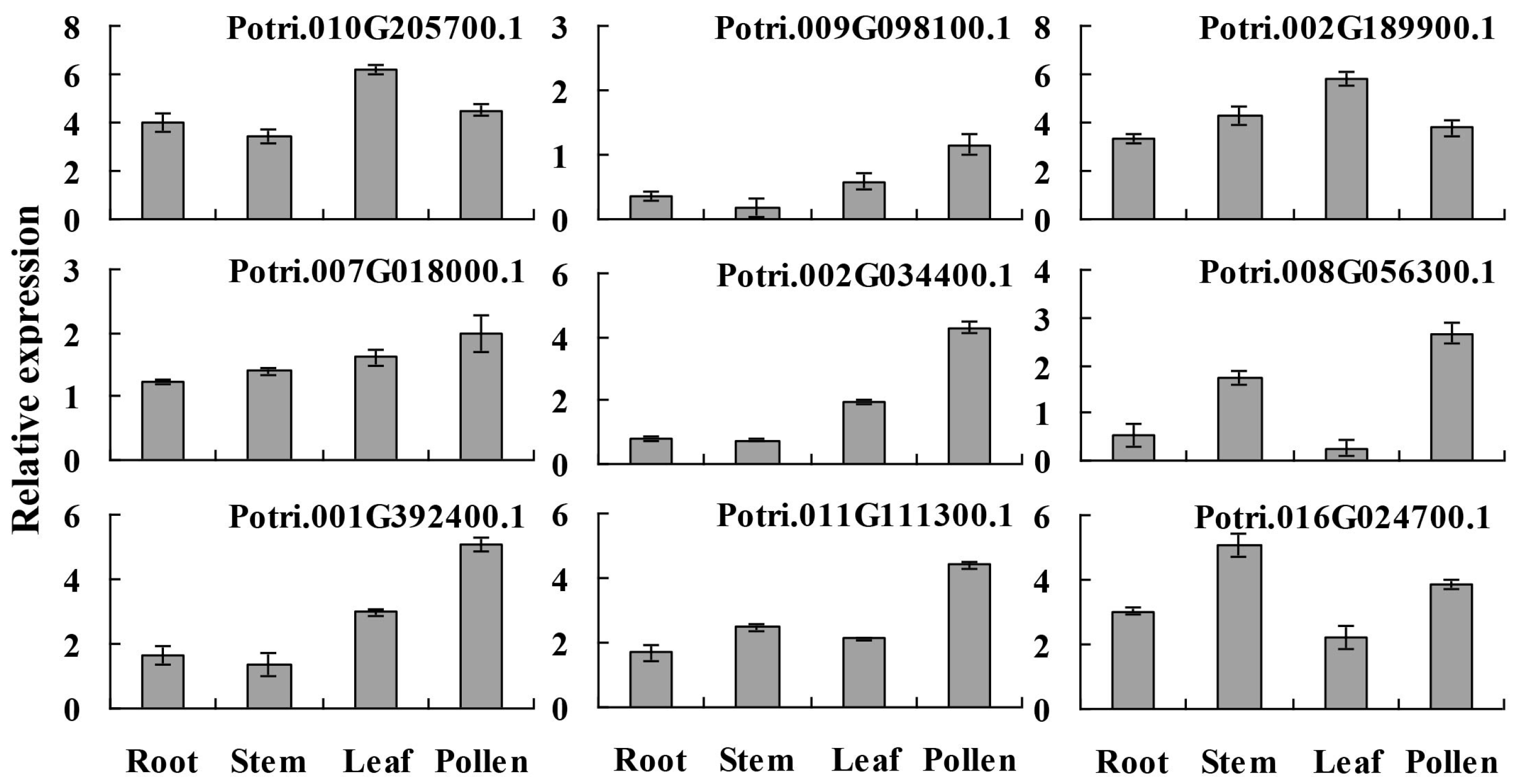
2.5. Expression Profiles of the Predicted Allergen Genes in Different Tissues
3. Materials and Methods
3.1. Plant Materials and Pollen Isolation
3.2. In Vitro Pollen Germination Assay
3.3. Preparation of Total Protein Extraction
3.4. Two-Dimensional Gel Electrophoresis (2-DE)
3.5. Gel Staining and Image Analysis
3.6. In-Gel Digestion and Mass Spectrometry
3.7. Protein Identification and Allergen Prediction
3.8. Microarray Data Analyses
3.9. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) ANalysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2-DE | Two-dimensional gel electrophoresis |
| ACN | Acetonitrile |
| CCB | Colloidal coomassie blue g-250 |
| CHAPS | 3-(3-cholamidopropyl) dimethylammonio-1-propane sulfonate |
| CHCA | α-cyano-4-hydroxycinnamic acid |
| DTT | Dithiothreitol |
| FDA | Fluorescein diacetate |
| GCRMA | Gene chip robust multiarray analysis |
| GEO | Gene expression omnibus |
| GO | Gene ontology |
| IEF | Isoelectric focusing |
| IPG | immobilized pH gradient |
| LC-MS/MS | Liquid chromatography coupled to tandem mass spectrometry |
| MALDI-TOF | Matrix-assisted laser desorption/ionization time-of-flight |
| MS | Mass spectrometry |
| MW | molecular weight |
| pI | isoelectric point |
| PMF | Peptide mass fingerprint |
| SDAP | Structural database of allergenic proteins |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TFA | Trifluoroacetic acid |
References
- Sheoran, I.S.; Sproule, K.A.; Olson, D.J.H.; Ross, A.R.S.; Sawhney, V.K. Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex. Plant Reprod. 2006, 19, 185–196. [Google Scholar] [CrossRef]
- Sheoran, I.S.; Ross, A.R.S.; Olson, D.J.H.; Sawhney, V.K. Proteomic analysis of tomato (Lycopersicon esculentum) pollen. J. Exp. Bot. 2007, 58, 3525–3535. [Google Scholar] [CrossRef] [PubMed]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Roulston, T.H.; Cane, J.H. Pollen nutritional content and digestibility for animals. Plant Syst. Evol. 2000, 222, 187–209. [Google Scholar] [CrossRef]
- AllergenOnline, the Farrp Allergen Protein Database. Available online: http://www.allergenonline.com/.
- Ivanciuc, O.; Schein, C.H.; Braun, W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 2003, 31, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Breiteneder, H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J. Allergy Clin. Immunol. 2006, 117, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Solomon, P.A. Air pollution and health: Bridging the gap from sources to health outcomes. Environ. Health Perspect. 2011, 119, 156. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Jahan, S.A.; Kabir, E. A review on human health perspective of air pollution with respect to allergies and asthma. Environ. Int. 2013, 59, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Li, L.; Chen, T.; Chong, K.; Xue, Y.; Wang, T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 2006, 6, 2504–2529. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Song, L.; Zhang, W.; Wang, Y.; Ruan, S.; Wu, W.H. Comparative proteomic analysis of Arabidopsis mature pollen and germinated pollen. J. Integr. Plant Biol. 2009, 51, 438–455. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Teshima, R. Proteomics-based allergen analysis in plants. J. Proteom. 2013, 93, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Mani, B.M.; Huerta-Ocampo, J.A.; Garcia-Sanchez, J.R.; Barrera-Pacheco, A.; de la Rosa, A.P.; Teran, L.M. Identification of Ligustrum lucidum pollen allergens using a proteomics approach. Biochem. Biophys. Res. Commun. 2015, 468, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Akagawa, M.; Handoyo, T.; Ishii, T.; Kumazawa, S.; Morita, N.; Suyama, K. Proteomic analysis of wheat flour allergens. J. Agric. Food Chem. 2007, 55, 6863–6870. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Sircar, G.; Pandey, N.; Bhattacharya, S.G. Mining novel allergens from coconut pollen employing manual de novo sequencing and homology-driven proteomics. J. Proteome Res. 2015, 14, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Pandey, N.; Bhattacharya, S.G. Charting novel allergens from date palm pollen (Phoenix sylvestris) using homology driven proteomics. J. Proteom. 2017, 165, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 2002, 90, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Zhang, J.; Guo, Y.H.; Sun, P.; Jia, H.X.; Fan, W.; Lu, M.Z.; Hu, J.J. Comparative proteomic analysis of mature pollen in triploid and diploid Populus deltoides. Int. J. Mol. Sci. 2016, 17, 1475. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Jensen, O.N. Modification-specific proteomics: Characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004, 8, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Holmes-Davis, R.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; McCormick, S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 2005, 5, 4864–4884. [Google Scholar] [CrossRef] [PubMed]
- Noir, S.; Bräutigam, A.; Colby, T.; Schmidt, J.; Panstruga, R. A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem. Biophys. Res. Commun. 2005, 337, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Brett, D.; Pospisil, H.; Valcárcel, J.; Reich, J.; Bork, P. Alternative splicing and genome complexity. Nat. Genet. 2002, 30, 29–30. [Google Scholar] [CrossRef] [PubMed]
- Loraine, A.E.; McCormick, S.; Estrada, A.; Patel, K.; Qin, P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013, 162, 1092–1109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, L.S.; Fan, W.; Zhang, X.L.; Jia, H.X.; Li, Y.; Yin, Y.F.; Hu, J.J.; Lu, M.Z. Proteomic analysis and candidate allergenic proteins in Populus deltoides CL. ‘2KEN8’ mature pollen. Front. Plant Sci. 2015, 6, 548. [Google Scholar] [CrossRef] [PubMed]
- Burbach, G.J.; Heinzerling, L.M.; Edenharter, G.; Bachert, C.; Bindslev-Jensen, C.; Bonini, S.; Bousquet, J.; Bousquet-Rouanet, L.; Bousquet, P.J.; Bresciani, M.; et al. GA2LEN skin test study II: Clinical relevance of inhalant allergen sensitizations in Europe. Allergy 2009, 64, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Sicherer, S.H. Clinical implications of cross-reactive food allergens. J. Allergy Clin. Immunol. 2001, 108, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Ortona, E.; Margutti, P.; Delunardo, F.; Vaccari, S.; Riganò, R.; Profumo, E.; Buttari, B.; Teggi, A.; Siracusano, A. Molecular and immunological characterization of the C-terminal region of a new Echinococcus granulosus Heat Shock Protein 70. Parasite Immunol. 2003, 25, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Chiung, Y.M.; Lin, B.L.; Yeh, C.H.; Lin, C.Y. Heat shock protein (hsp 70)-related epitopes are common allergenic determinants for barley and corn antigens. Electrophoresis 2000, 21, 297–300. [Google Scholar] [CrossRef]
- Shen, H.D.; Au, L.C.; Lin, W.L.; Liaw, S.F.; Tsai, J.J.; Han, S.H. Molecular cloning and expression of a Penicillium citrinum allergen with sequence homology and antigenic crossreactivity to a hsp 70 human heat shock protein. Clin. Exp. Allergy 1997, 27, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, C.; Poysa, V.; Cober, E.R.; Gleddie, S. Soybean allergens affecting North American patients identified by 2D gels and mass spectrometry. Food Anal. Methods 2010, 3, 363–374. [Google Scholar] [CrossRef]
- Simon-Nobbe, B.; Probst, G.; Kajava, A.V.; Oberkofler, H.; Susani, M.; Crameri, R.; Ferreira, F.; Ebner, C.; Breitenbach, M. IgE-binding epitopes of enolases, a class of highly conserved fungal allergens. J. Allergy Clin. Immunol. 2000, 106, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Gupta, R.; Jhingran, A.; Singh, B.P.; Sridhara, S.; Gaur, S.N.; Arora, N. Cloning, recombinant expression and activity studies of a major allergen “enolase” from the fungus Curvularia lunata. J. Clin. Immunol. 2006, 26, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Alché, J.D.; M’rani-Alaoui, M.; Castro, A.J.; Rodríguez-García, M.I. Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 2004, 45, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Weichel, M.; Glaser, A.G.; Ballmer-Weber, B.K.; Schmid-Grendelmeier, P.; Crameri, R. Wheat and maize thioredoxins: A novel cross-reactive cereal allergen family related to baker’s asthma. J. Allergy Clin. Immunol. 2006, 117, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Van Ree, R.; Voitenko, V.; van Leeuwen, W.A.; Aalberse, R.C. Profilin is a cross-reactive allergen in pollen and vegetable foods. Int. Arch. Allergy Immunol. 1992, 98, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Asero, R.; Jimeno, L.; Barber, D. Preliminary results of a skin prick test-based study of the prevalence and clinical impact of hypersensitivity to pollen panallergens (polcalcin and profilin). J. Investig. Allergol. Clin. Immunol. 2010, 20, 35–38. [Google Scholar] [PubMed]
- Alvarado, M.I.; Jimeno, L.; De La Torre, F.; Boissy, P.; Rivas, B.; Lázaro, M.J.; Barber, D. Profilin as a severe food allergen in allergic patients overexposed to grass pollen. Allergy 2014, 69, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Sander, I.; Flagge, A.; Merget, R.; Halder, T.M.; Meyer, H.E.; Baur, X. Identification of wheat flour allergens by means of 2-dimensional immunoblotting. J. Allergy Clin. Immunol. 2001, 107, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.Z.; Song, L.F.; Zou, J.J.; Su, Z.; Wu, W.H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mascot Server, MASCOT Search Engine. Available online: http://www.matrixscience.com.
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 22 March 2012).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
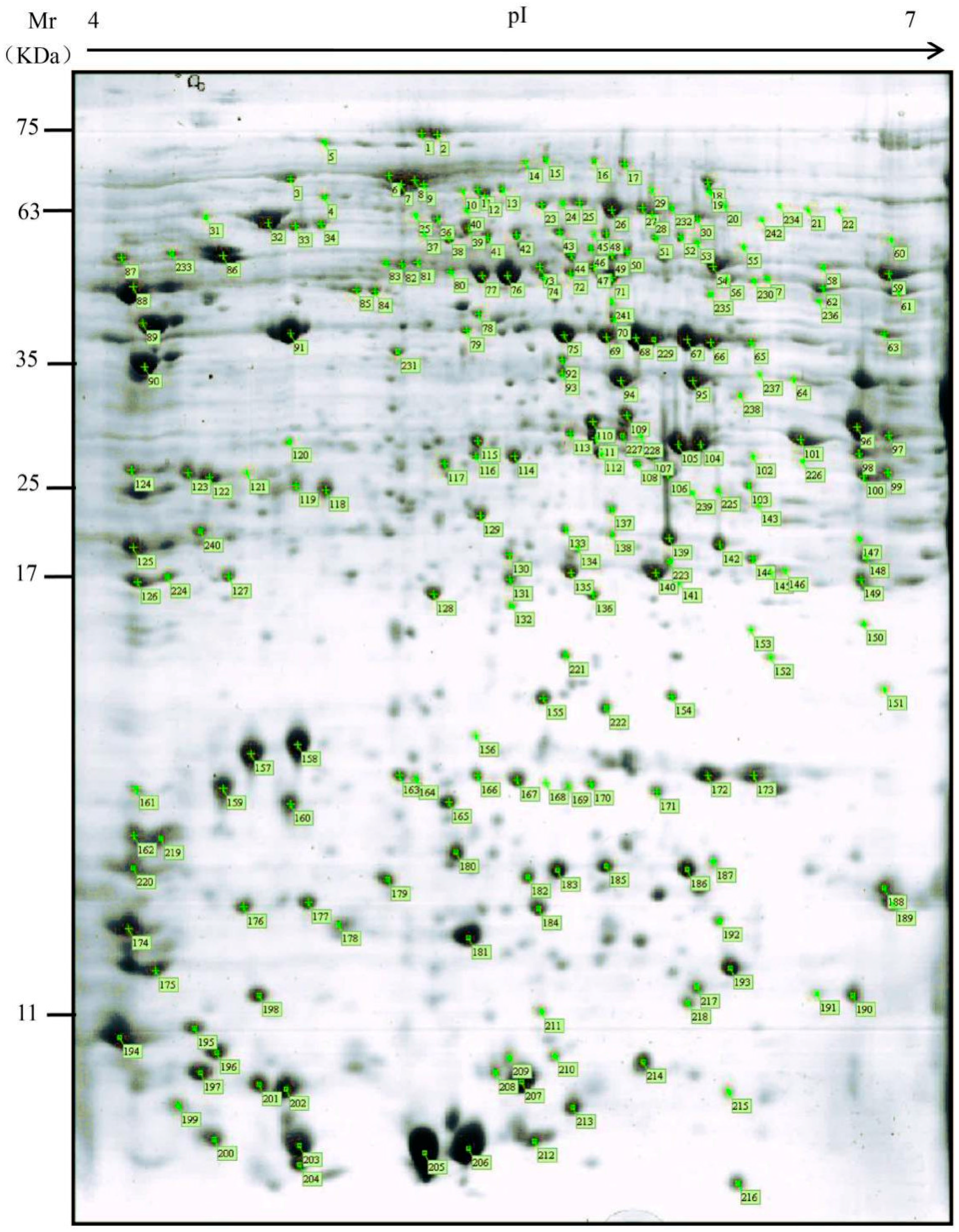


| Spot No. | Gene ID | Protein Description | Amino Acids (aa) | Corresponding of Known Allergen | ||||
|---|---|---|---|---|---|---|---|---|
| Allergen | Accession No. | Amino Acids (aa) | Bit Score | E Score | ||||
| 4/8/21 | Potri.003G143600.1 | Heat shock protein 70 (Hsp 70) family protein | 666 | Cor a 10 | CAC14168 | 668 | 791.1 | 0.0 |
| 5/7 | Potri.003G006300.1 | Chloroplast heat shock protein 70-2 | 706 | Cor a 10 | CAC14168 | 668 | 322.5 | 5.0 × 10−89 |
| 9 | Potri.010G205700.1 | Heat shock protein 70 (Hsp 70) family protein | 648 | Cla h 5.0101 | P40918 | 643 | 605.2 | 3.7 × 10−174 |
| 10/12/13 | Potri.009G079700.1 | Mitochondrial HSO70 2 | 682 | Cor a 10 | CAC14168 | 668 | 392.8 | 3.4 × 10−110 |
| 11 | Potri.001G285500.1 | Mitochondrial HSO70 2 | 683 | Cor a 10 | CAC14168 | 668 | 400.4 | 1.7 × 10−112 |
| 25/62/71/72/241 | Potri.006G116800.1 | Enolase | 445 | Hev b 9 | Q9LEI9 | 445 | 583.5 | 5.6 × 10−168 |
| 31 | Potri.001G087500.1 | Heat shock protein 70 (Hsp 70) family protein | 666 | Cor a 10 | CAC14168 | 668 | 816.5 | 0.0 |
| 43 | Potri.009G098100.1 | Granulin repeat cysteine protease family protein | 508 | Act d 1 | AAA32629 | 380 | 237.7 | 7.0 × 10−64 |
| 48 | Potri.002G189900.1 | Aldehyde dehydrogenase 2B7 | 540 | Cla h 10.0101 | P40108 | 496 | 364.1 | 8.8 × 10−102 |
| 49/50/53/54/59/222 | Potri.015G131100.1 | Enolase | 445 | Hev b 9 | Q9LEJ0 | 445 | 545.7 | 1.4 × 10−156 |
| 55 | Potri.007G018000.1 | Thioredoxin H-type 1 | 122 | Tri a 25.0101 | Q9LDX4 | 125 | 114.8 | 5.5 × 10−28 |
| 88 | Potri.005G015100.1 | Calreticulin 1a | 419 | Pen ch 31.0101 | AAX45072 | 557 | 91.6 | 7.9 × 10−20 |
| 90 | Potri.013G009500.1 | Calreticulin 1a | 360 | Pen ch 31.0101 | AAX45072 | 557 | 90.2 | 1.9 × 10−19 |
| 98/105 | Potri.002G034400.1 | NmrA-like negative transcriptional regulator family protein | 308 | Bet v 6.0102 | AAG22740 | 308 | 353.3 | 5.3 × 10−99 |
| 106 | Potri.017G102000.1 | Malate dehydrogenase | 412 | Mala f 4 | AAD25927 | 342 | 244.0 | 6.4 × 10−66 |
| 122/123 | Potri.012G090900.1 | Cysteine proteinases superfamily protein | 363 | Act d 1 | AAA32629 | 380 | 238.2 | 3.6 × 10−64 |
| 145 | Potri.008G056300.1 | Triosephosphate isomerase | 255 | Tri a 31.0101 | CAC14917 | 253 | 332.0 | 9.6 × 10−93 |
| 156 | Potri.001G392400.1 | Pollen Ole e 1 allergen and extensin family protein | 161 | Lyc e LAT52 | CAA33854 | 161 | 118.4 | 7.8 × 10−29 |
| 159/160 | Potri.011G111300.1 | Pollen Ole e 1 allergen and extensin family protein | 164 | Lyc e LAT52 | CAA33854 | 161 | 120.8 | 1.5 × 10−29 |
| 172/173 | Potri.013G089200.1 | HSP20-like chaperones superfamily protein | 192 | Cas s 9.0101 | CAE46905 | 154 | 103.8 | 2.1 × 10−24 |
| 183 | Potri.018G083500.1 | Thioredoxin-dependent peroxidase 1 | 162 | Cand b 2 | AAA34357 | 167 | 105.4 | 6.6 × 10−25 |
| 194 | Potri.016G024700.1 | Calmodulin 6 | 149 | Tyr p 24.0101 | ACL36923 | 153 | 112.0 | 5.8 × 10−27 |
| 197 | Potri.003G047700.1 | Profilin 3 | 131 | Hev b 8.0201 | Q9M7N0 | 131 | 195.6 | 2.9 × 10−52 |
| 202 | Potri.001G190800.1 | Profilin 5 | 133 | Hev b 8.0201 | Q9M7N0 | 131 | 144.2 | 9.0 × 10−37 |
| 210 | Potri.005G232700.1 | Thioredoxin H-type 1 | 114 | Tri a 25.0101 | Q9LDX4 | 125 | 115.4 | 3.3 × 10−28 |
| 215 | Potri.009G022300.1 | Cystatin B | 100 | Act d 4.0101 | AAR92223 | 116 | 69.1 | 2.4 × 10−14 |
| 217/218 | Potri.006G093500.1 | HSP20-like chaperones superfamily protein | 140 | Cas s 9.0101 | CAE46905 | 154 | 151.4 | 7.6 × 10−39 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhang, X.; Zhang, J.; Fan, W.; Lu, M.; Hu, J. Proteomic Analysis and Identification of Possible Allergenic Proteins in Mature Pollen of Populus tomentosa. Int. J. Mol. Sci. 2018, 19, 250. https://doi.org/10.3390/ijms19010250
Wang L, Zhang X, Zhang J, Fan W, Lu M, Hu J. Proteomic Analysis and Identification of Possible Allergenic Proteins in Mature Pollen of Populus tomentosa. International Journal of Molecular Sciences. 2018; 19(1):250. https://doi.org/10.3390/ijms19010250
Chicago/Turabian StyleWang, Liuqiang, Xiaoling Zhang, Jin Zhang, Wei Fan, Mengzhu Lu, and Jianjun Hu. 2018. "Proteomic Analysis and Identification of Possible Allergenic Proteins in Mature Pollen of Populus tomentosa" International Journal of Molecular Sciences 19, no. 1: 250. https://doi.org/10.3390/ijms19010250





