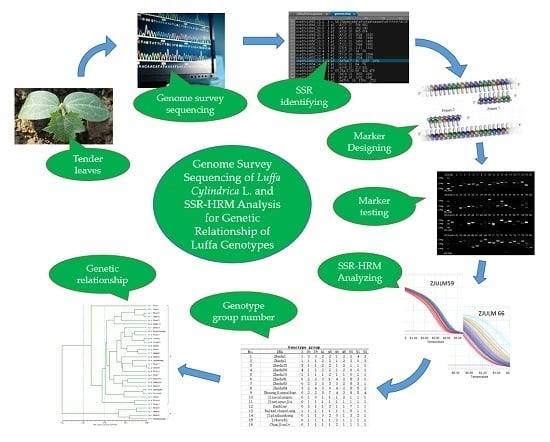
Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes
Abstract
:1. Introduction
2. Results
2.1. Genome Sequencing and Sequence Assembly
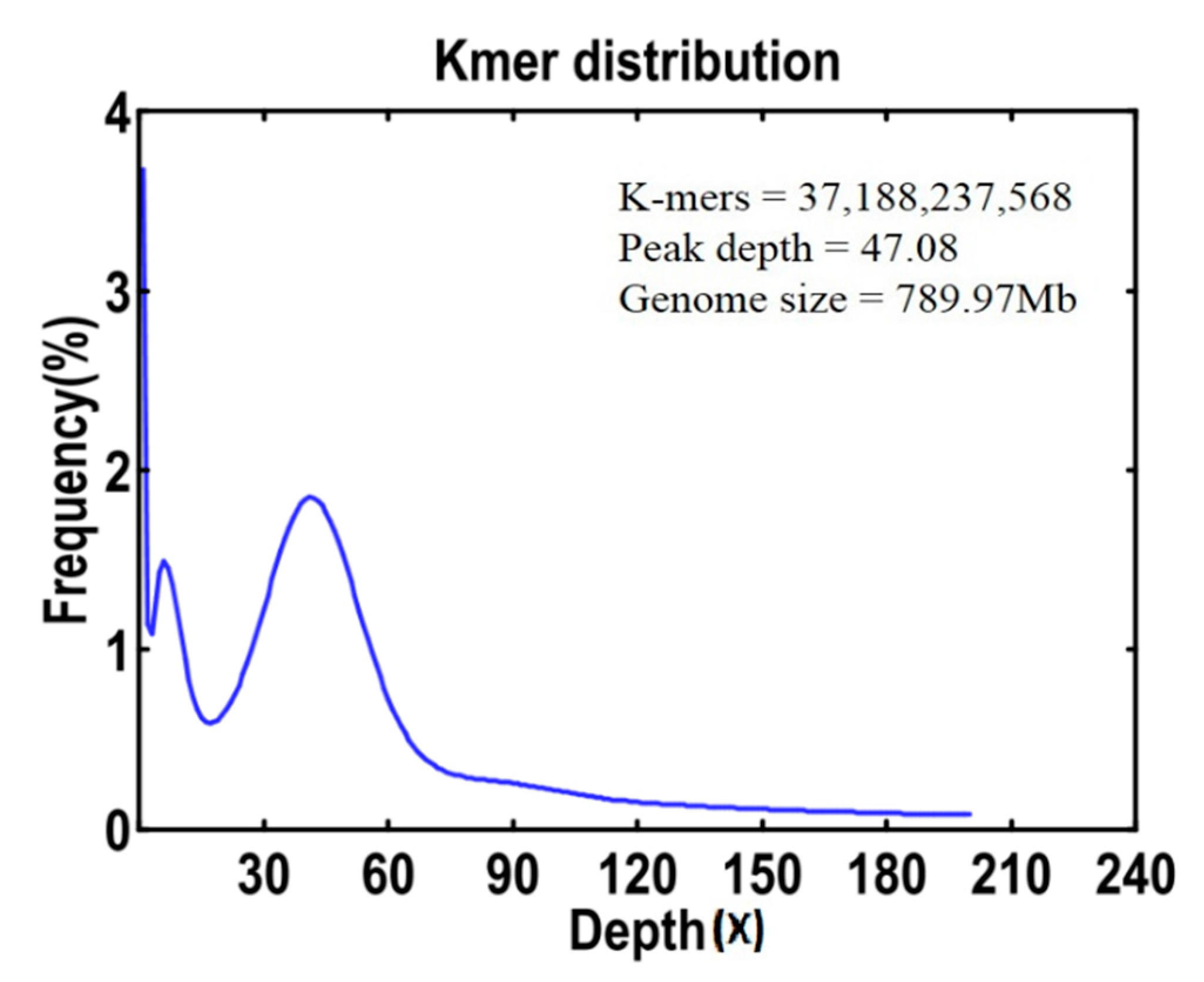
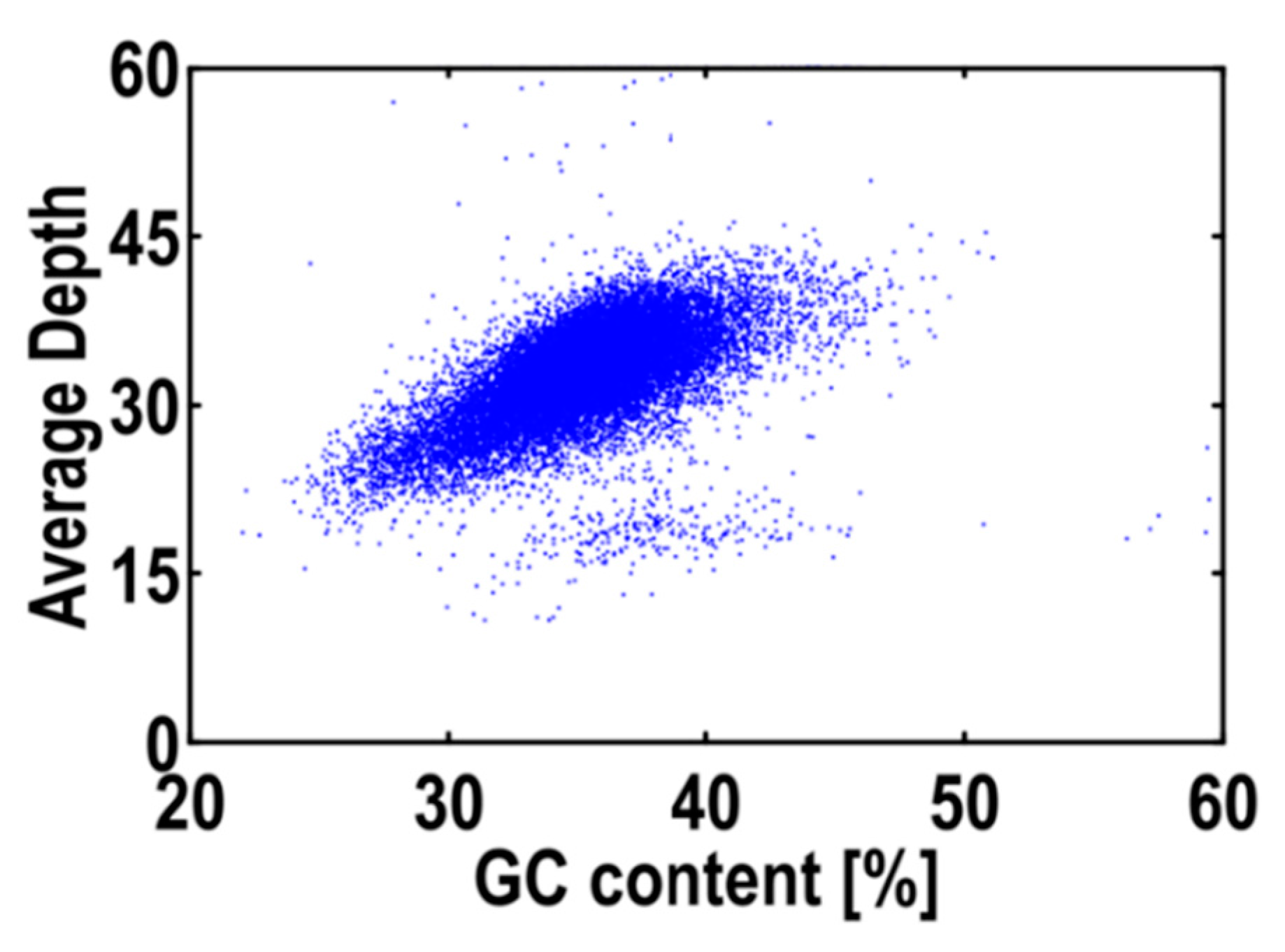
2.2. Genome Size Estimation, GC Content and Genome Survey
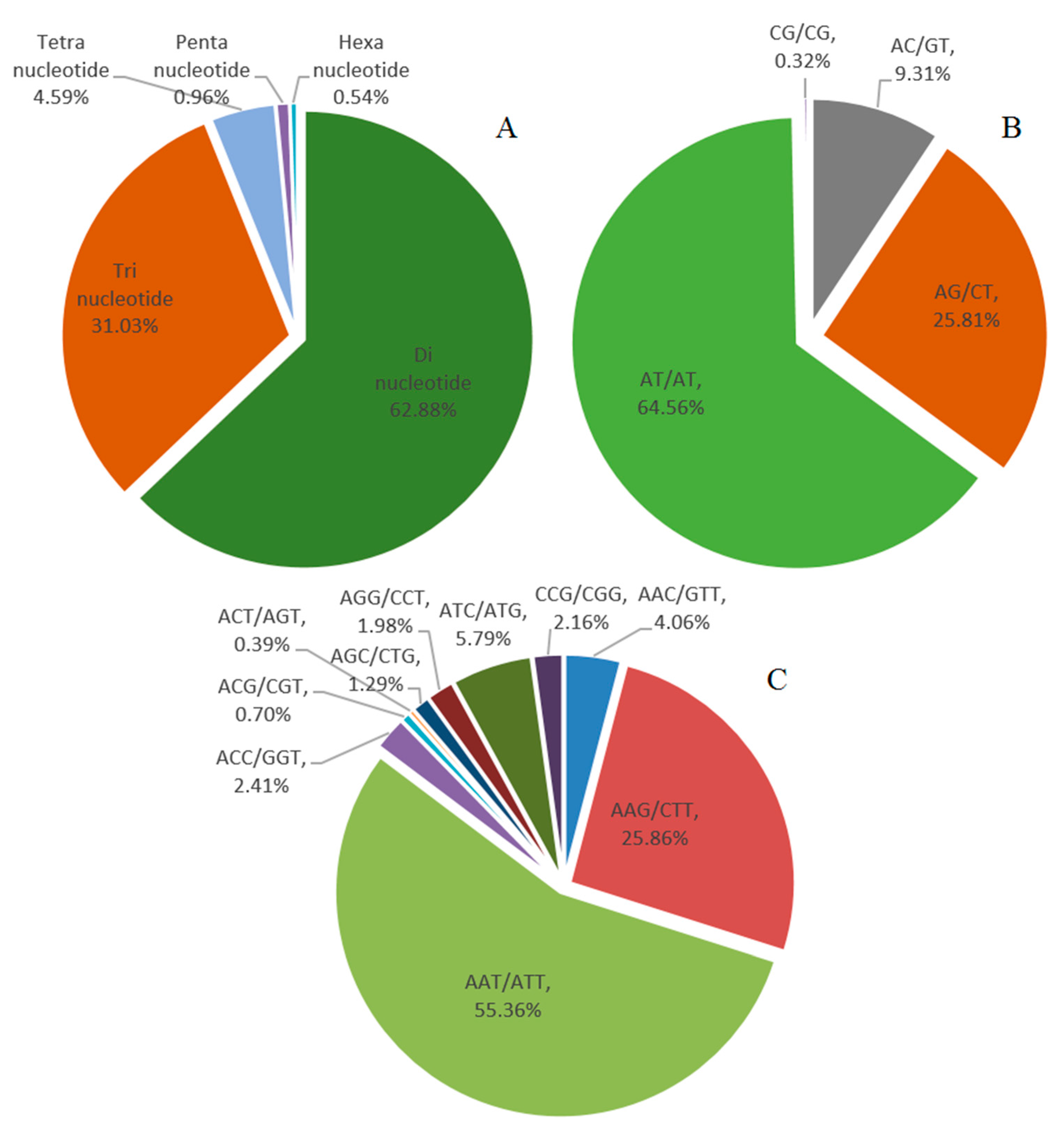
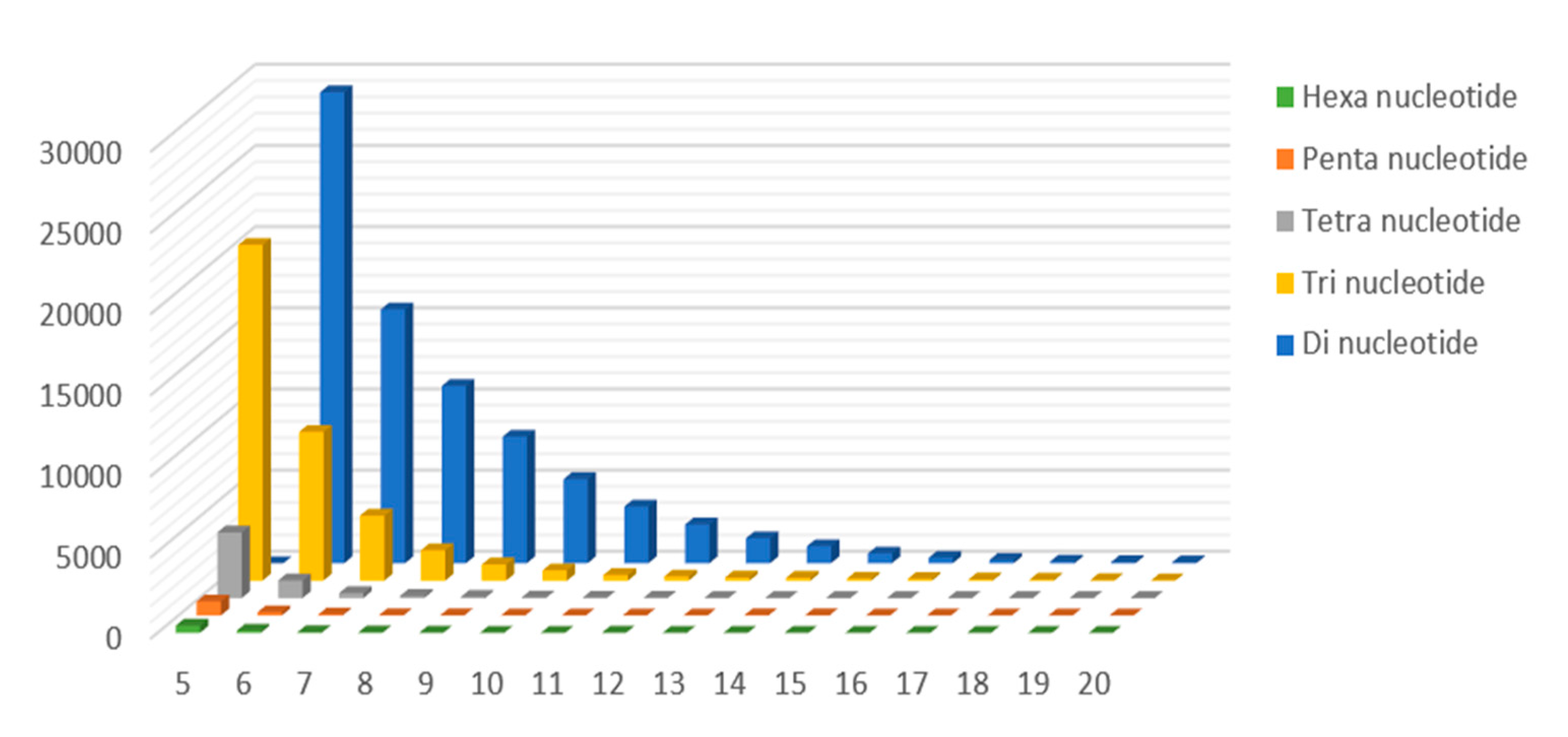
2.3. Genomic SSR Markers Development
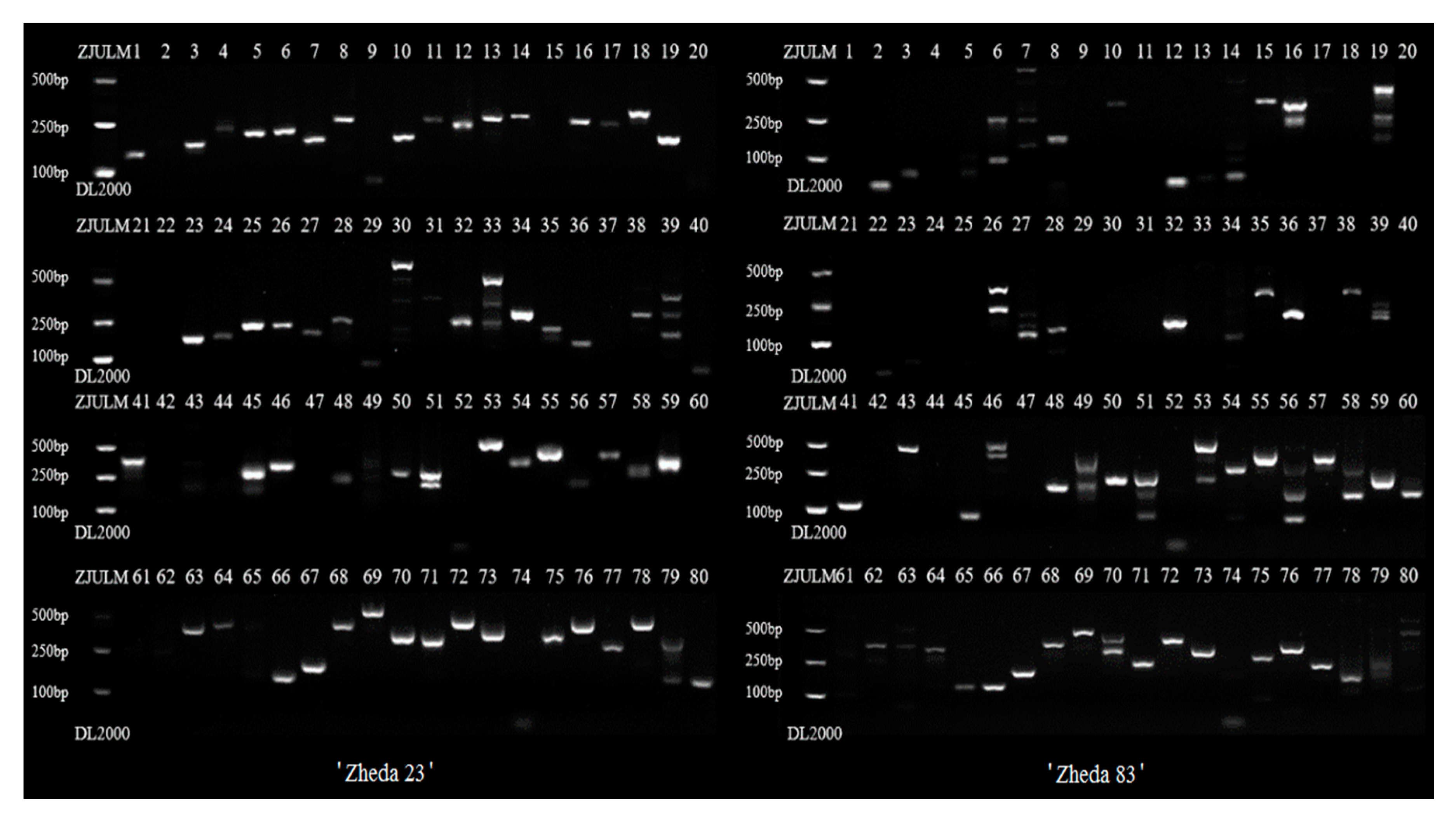
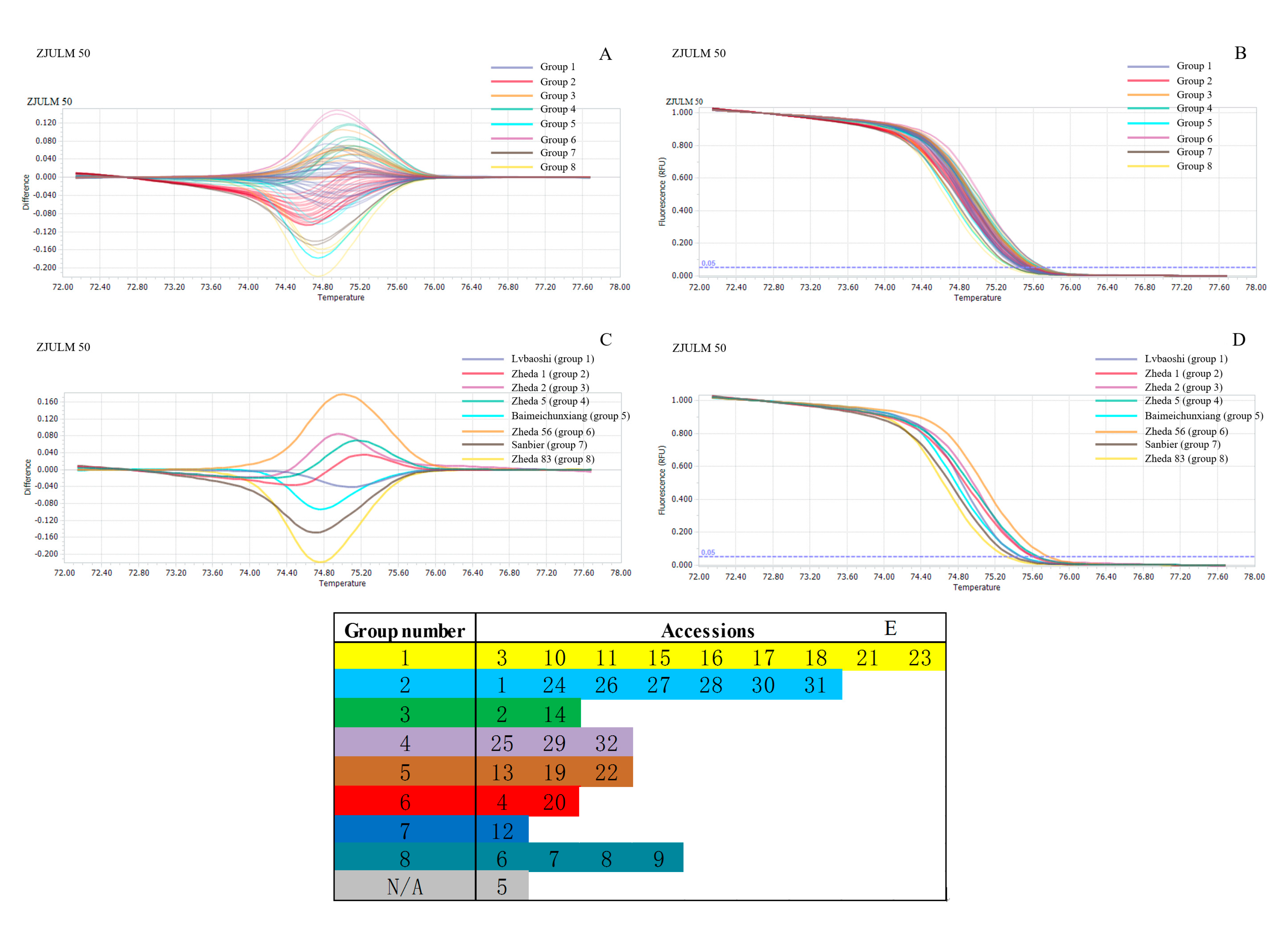
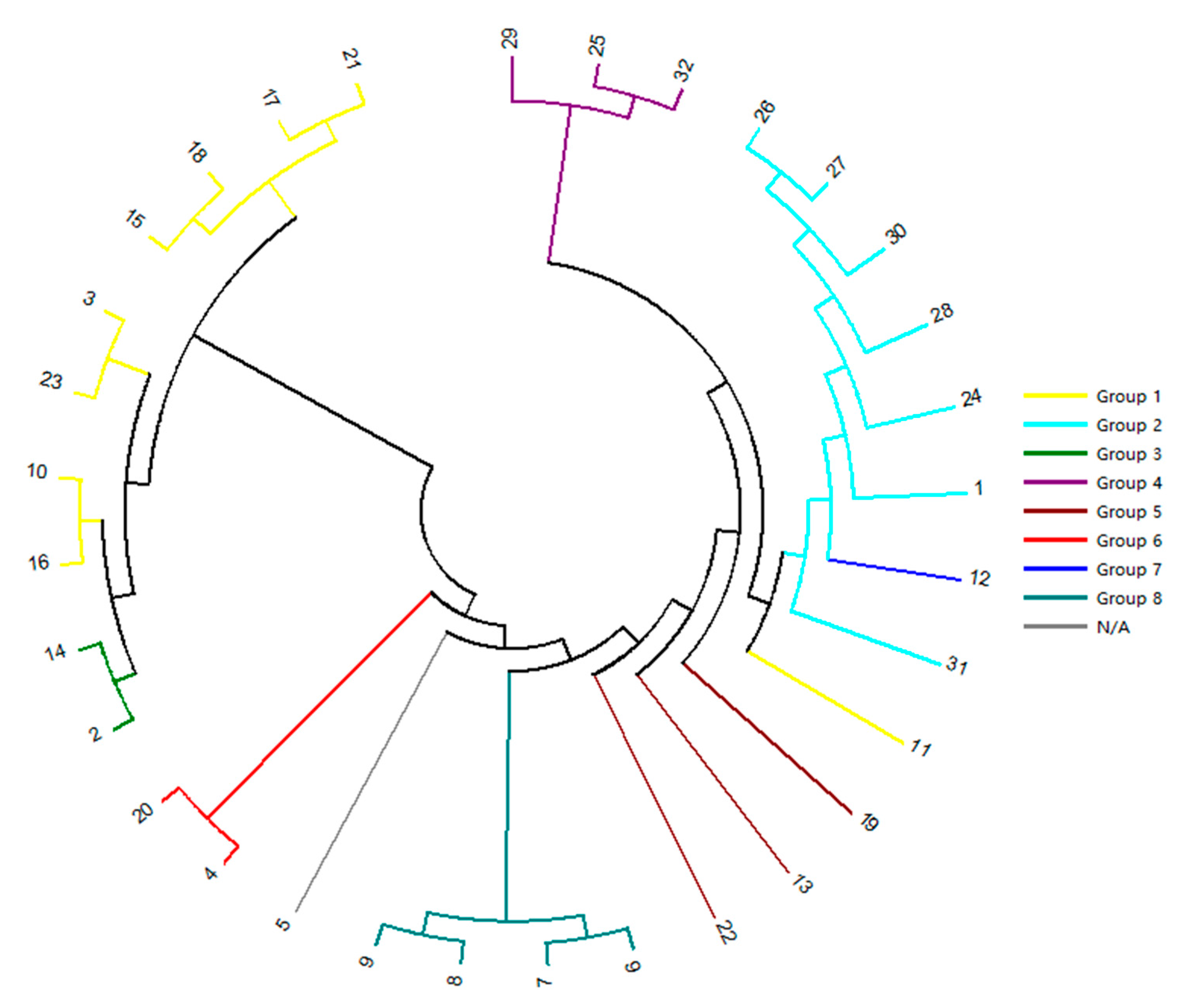
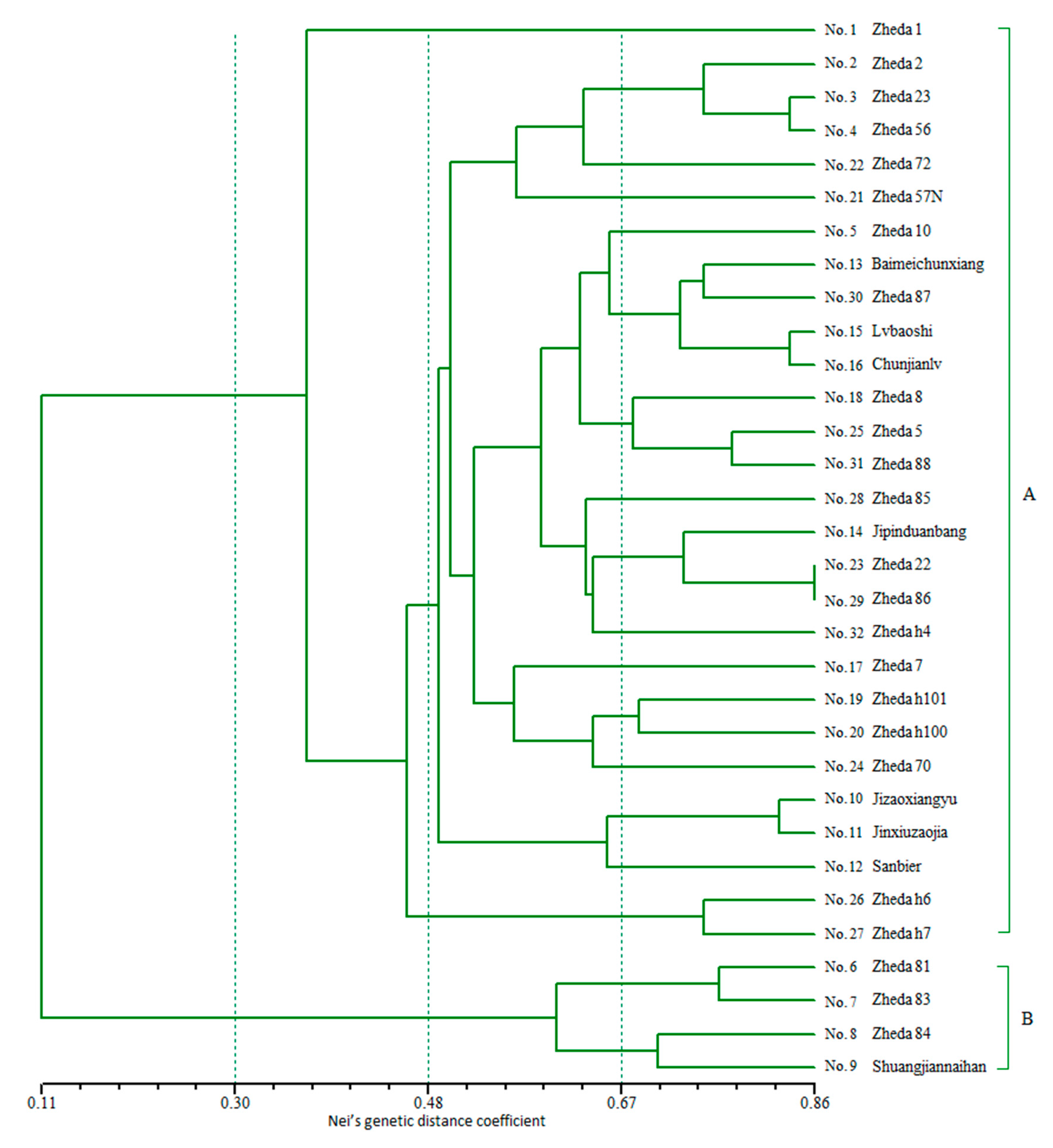
2.4. Genetic Relationship Analysis by SSR-HRM
3. Discussion
3.1. Characteristics of Luffa cylindrica L. Genome
3.2. Genomic SSR Markers Development
3.3. Genetic Relationship Analysis by SSR-HRM
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. Genome Sequencing and Sequence Assembly
4.3. Genome Size Estimation, GC Content and Genome Survey
4.4. Genomic SSR Marker Development
4.5. Genetic Relationship Analysis by SSR-HRM
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SSR-HRM | Microsatellite high resolution melting |
| NGS | Next-generation sequencing |
| SSR | Microsatellite |
| UPGMA | Unweighted pair group method analysis |
| EST | Expressed sequence tag |
| ZJULM | Zhejiang University luffa marker |
| MISA | Microsatellite searching tool |
| GC | Guanine pluscytosine |
| NCBI | National center for biotechnology information |
| HRM | High resolution melting |
| PCR | Polymerase chain reaction |
| MEGA | Molecular evolutionary genetics analysis |
| PIC | Polymorphism information content |
| PAGE | Polyacrylamide gel electrophoresis |
References
- Wu, H.B.; He, X.L.; Gong, H.; Luo, S.B.; Li, M.Z.; Chen, J.Q.; Zhang, C.Y.; Yu, T.; Huang, W.P.; Luo, J.N. Genetic linkage map construction and QTL analysis of two interspecific reproductive isolation traits in sponge gourd. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.K.; Kc, H.B.; Tiwari, R.K.; Ghale, M.; Sthapit, B.R. Descriptors for sponge gourd (Luffa cylindrica (L.) Roem.). Narc Kathmandu Np; 2004. Available online: https://idl-bnc-idrc.dspacedirect.org/bitstream/handle/10625/31459/122785.pdf?sequence=1.
- Partap, S.; Kumar, A.; Sharma, N.K.; Jha, K.K. Luffa cylindrica: An important medicinal plant. J. Nat. Prod. Plant Resour. 2012, 2, 127–134. [Google Scholar]
- Sheng, Z.; Jin, H.; Zhang, C.F.; Guan, Y.J.; Ying, Z. Genetic analysis of fruit shape traits at different maturation stages in sponge gourd. J. Zhejiang Univ. Sci. B 2007, 8, 338–344. [Google Scholar]
- Lu, M.; An, H.M.; Li, L.L. Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii tratt and leaf ascorbate metabolism genes. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hu, Y.Y.; Sui, Z.H.; Fu, F.; Wang, J.G.; Chang, L.P.; Guo, W.H.; Li, B.B. Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis (Rhodophyta) based on next-generation sequencing. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Jia, H.M.; Li, X.W.; Chai, M.L.; Jia, H.J.; Chen, Z.; Wang, G.Y.; Chai, C.Y.; van de Weg, E.; Gao, Z.S. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.B.; Gong, H.; Liu, P.; He, X.L.; Luo, S.B.; Zheng, X.M.; Zhang, C.Y.; He, X.M.; Luo, J.N. Large-scale development of EST-SSR markers in sponge gourd via transcriptome sequencing. Mol. Breed. 2014, 34, 1903–1915. [Google Scholar] [CrossRef]
- Wang, Y.W.; Samuels, T.D.; Wu, Y.Q. Development of 1,030 genomic SSR markers in switchgrass. Theor. Appl. Genet. 2011, 122, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, J.; Pingoud, A.; Hahn, M. Validation of an algorithm for automatic quantification of nucleic acid copy numbers by real-time polymerase chain reaction. Anal. Biochem. 2003, 317, 218–225. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Wittwer, C.T. High-resolution DNA melting analysis: Advancements and limitations. Hum. Mutat. 2009, 30, 857–859. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Reed, G.H.; Gundry, C.N.; Vandersteen, J.G.; Pryor, R.J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003, 49, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Tindall, E.A.; Petersen, D.C.; Woodbridge, P.; Schipany, K.; Hayes, V.M. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum. Mutat. 2009, 30, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Bosmali, I.; Ganopoulos, I.; Madesis, P.; Tsaftaris, A. Microsatellite and DNA-barcode regions typing combined with high resolution melting (HRM) analysis for food forensic uses: A case study on lentils (lens culinaris). Food Res. Int. 2012, 46, 141–147. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Ganopoulos, I.; Koubouris, G.; Tsaftaris, A.; Sergendani, C.; Kalivas, A.; Madesis, P. Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europaea germplasm collection. Plant Genet. Resour. 2014, 12, 273–277. [Google Scholar] [CrossRef]
- Chitsaz, H.; Yee-Greenbaum, J.L.; Tesler, G.; Lombardo, M.J.; Dupont, C.L.; Badger, J.H.; Novotny, M.; Rusch, D.B.; Fraser, L.J.; Gormley, N.A.; et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat. Biotechnol. 2011, 29, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method—A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; Gonzalez, V.M.; Henaff, E.; Camara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.W.; Xie, B.Y.; Ni, P.X.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.Y.; Sun, Z.X.; Bin, L.H.; Xu, D.M.; Wu, B.; Zhang, B.; Wang, X.C.; Han, Y.H.; Zhang, L.J.; Qiao, Z.J.; et al. Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Mol. Biol. Rep. 2016, 34, 233–241. [Google Scholar] [CrossRef]
- Cheung, M.S.; Down, T.A.; Latorre, I.; Ahringer, J. Systematic bias in high-throughput sequencing data and its correction by beads. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in illumina sequencing libraries. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Okada, Y.; Tabuchi, H.; Shirasawa, K.; Watanabe, A.; Tsuruoka, H.; Minami, C.; Nakayama, S.; Sasamoto, S.; Kohara, M.; et al. Survey of genome sequences in a wild sweet potato, Ipomoea trifida (H.B.K.) G. Don. DNA Res. 2015, 22, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pan, S.K.; Cheng, S.F.; Zhang, B.; Mu, D.S.; Ni, P.X.; Zhang, G.Y.; Yang, S.; Li, R.Q.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Richards, S.; Desjardins, C.A.; Niehuis, O.; Gadau, J.; Colbourne, J.K.; Beukeboom, L.W.; Desplan, C.; Elsik, C.G.; Grimmelikhuijzen, C.J.P.; et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 2010, 327, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.S.; Yang, L.; Peng, Z.H.; Sun, H.Y.; Yue, X.H.; Lou, Y.F.; Dong, L.L.; Wang, L.L.; Gao, Z.M. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.Q.; Huang, S.M.; Zhan, J.P.; Yu, J.Y.; Wang, X.F.; Hua, W.; Liu, S.Y.; Liu, G.H.; Wang, H.H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 2014, 21, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Dong, Y.; Zhao, J.J.; Huang, L.; Ren, X.P.; Chen, Y.N.; Huang, S.M.; Liao, B.S.; Lei, Y.; Yan, L.Y.; et al. Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.). BMC Genomics 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Bassam, B.J.; Caetanoanolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence-limits on phylogenies—An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
Sample Availability: Genome survey sequencing data of Luffa cylindrica L. are available from the authors. |

| Library | Data (Gb) | Depth (×) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|
| 220 bp | 43.40 | 54.94 | 96.77 | 91.56 |
| Scaffold Number | Scaffold Length (bp) | Scaffold N50 (bp) | Scaffold N90 (bp) | Gap total Length (bp) |
| 1,410,117 | 885,010,283 | 807 | 266 | 9,841,708 |
| Contig Number | Contig Length (bp) | Contig N50 (bp) | Contig N90 (bp) | GC Content (%) |
| 1,913,731 | 875,168,575 | 525 | 236 | 34.34 |
| Searching Item | Number | Percentage |
|---|---|---|
| Total number of sequences examined | 2,697,125 | – |
| Total size of examined sequences (bp) | 1,064,011,890 | – |
| Total number of identified SSRs | 431,234 | 100.00% |
| Number of SSRs containing sequences | 218,940 | 50.77% |
| Number of sequences containing more than 1 SSR | 74,313 | 17.23% |
| Number of SSRs present in compound formation | 47,595 | 11.04% |
| Mono-nucleotide | 306,140 | 70.99% |
| Di-nucleotide | 78,655 | 18.24% |
| Tri-nucleotide | 38,823 | 9.00% |
| Tetra-nucleotide | 5737 | 1.33% |
| Penta-nucleotide | 1196 | 0.28% |
| Hexa-nucleotide | 683 | 0.16% |
| Locus | GenBank Accession | Repeat Motif | Primer Sequence (5′–3′) | Gene Diversity | PIC |
|---|---|---|---|---|---|
| ZJULM 3 | KY983987 | (TTTTAT)6 | Forward: CGTGTCTTCGGGATAATA Reverse: CGAAGCATCTTTTACCAT | 0.6032 | 0.5556 |
| ZJULM 36 | KY983988 | (CTTCCA)6 | Forward: CTGAGCAGTCTAACACCCAT Reverse: TGTGCGAACAAGGAAGGA | 0.4141 | 0.3874 |
| ZJULM 39 | KY983989 | (AAAAAT)5 | Forward: GCGAGTATAGCTCAACGG Reverse: TTCCAAAATCCAAACCAA | 0.3673 | 0.3484 |
| ZJULM 41 | KY983990 | (TGATGG)5 | Forward: TCGTTGGTGTTGTAGGGTTT Reverse: GAGGACGAATTGGAAGGAGT | 0.6900 | 0.6404 |
| ZJULM 45 | KY983991 | (TTTTA)5 | Forward: AATTCCCAGGTAATGTTATG Reverse: GTCGGCTTGTTTCTTCTC | 0.6563 | 0.5888 |
| ZJULM 46 | KY983992 | (AAGAG)5 | Forward: CCCGCAGTGTTAAGTTTC Reverse: CCTGCCATGTTTGTTCTC | 0.4019 | 0.3756 |
| ZJULM 48 | KY983993 | (AAAAT)5 | Forward: TAGAAAGGAAAGGAGGAA Reverse: TTCAAGAGTTCAGGGTTT | 0.6240 | 0.5527 |
| ZJULM 50 | KY983994 | (TAAGA)5 | Forward: TTCTCCAAATAAGCCACT Reverse: AGAATCTCCTACCCGTTT | 0.8137 | 0.7896 |
| ZJULM 51 | KY983995 | (TGGTTG)6 | Forward: CCAGTCCAGGAGAAAGGG Reverse: GAGGCACAACCACAACCA | 0.6505 | 0.6129 |
| ZJULM 53 | KY983996 | (GAAAGT)5 | Forward: GCGAAGAGGAGCGAAGAA Reverse: ATTGGCAATGCAATGAGG | 0.3889 | 0.3613 |
| ZJULM 55 | KY983997 | (CCAGCA)5 | Forward: GATAATGGAAATAAACCACCCT Reverse: GCCACAGACCCTACTTGAGA | 0.7813 | 0.7506 |
| ZJULM 59 | KY983998 | (CCACCT)7 | Forward: CGTGTAGGCTAGGGTCAC Reverse: CTCCACCACTTCATTTGTAT | 0.5898 | 0.5418 |
| ZJULM 66 | KY983999 | (TTTTGT)5 | Forward: AAGATCGGTTTGGGAGGA Reverse: TGGCAGTTTCAGGCAGTC | 0.6468 | 0.6156 |
| ZJULM 67 | KY984000 | (GTTTTT)5 | Forward: GGGATATTGCGGTGGAGT Reverse: GGTTAGGTGGCGTTCGTC | 0.7111 | 0.6637 |
| ZJULM 71 | KY984001 | (ATTTTT)5 | Forward: AGTTCCCTGAGCAGATAC Reverse: CTAAATCAACAACATCCCT | 0.5729 | 0.5439 |
| ZJULM 73 | KY984002 | (TGGTTG)6 | Forward: TTGAGCCTGAGGGATAGA Reverse: GGATGCTGCTGATAAGTG | 0.7041 | 0.6798 |
| ZJULM 77 | KY984003 | (AGAGAA)5 | Forward: TTCGGTCATTTGATTTCG Reverse: TTCGTGGAAGAACCCTCT | 0.1913 | 0.1730 |
| ZJULM 78 | KY984004 | (AGTTCC)5 | Forward: GAACATCCCAGGAAATGC Reverse: GCCAGACGAGGAAGAACA | 0.2246 | 0.2096 |
| ZJULM 79 | KY984005 | (GAAAAA)5 | Forward: GAGGAGATGGTGAGGGAG Reverse: AACGGATTGCTGATGTGA | 0.6806 | 0.6427 |
| Mean | – | – | – | 0.5638 | 0.5281 |
| Accession Number | ZJU Luffa Marker | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 36 | 39 | 41 | 45 | 46 | 48 | 50 | 51 | 53 | 55 | 59 | 66 | 67 | 71 | 73 | 77 | 78 | 79 | |
| 1 | 1 | 3 | 0 | 2 | 2 | 1 | 2 | 2 | 4 | 3 | 6 | 2 | 3 | 3 | 1 | 0 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 3 | 0 | 3 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| 3 | 3 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 5 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| 4 | 4 | 1 | 1 | 2 | 2 | 1 | 2 | 6 | 0 | 1 | 5 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| 5 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 6 | 1 | 3 | 1 | 1 | 5 | 1 | 1 | 1 |
| 6 | 5 | 2 | 2 | 3 | 1 | 4 | 2 | 8 | 3 | 1 | 2 | 3 | 5 | 4 | 2 | 7 | 1 | 2 | 4 |
| 7 | 0 | 2 | 2 | 3 | 3 | 3 | 2 | 8 | 3 | 1 | 2 | 3 | 4 | 4 | 2 | 0 | 1 | 3 | 4 |
| 8 | 3 | 2 | 4 | 3 | 0 | 4 | 2 | 8 | 5 | 2 | 2 | 3 | 4 | 5 | 2 | 8 | 1 | 2 | 5 |
| 9 | 0 | 2 | 2 | 3 | 4 | 4 | 2 | 8 | 5 | 4 | 2 | 3 | 5 | 4 | 2 | 1 | 1 | 2 | 5 |
| 10 | 0 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 1 | 6 | 1 | 1 | 3 |
| 11 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 2 | 1 | 3 | 1 | 6 | 1 | 1 | 3 |
| 12 | 0 | 3 | 1 | 1 | 2 | 1 | 1 | 7 | 1 | 1 | 1 | 2 | 6 | 3 | 1 | 1 | 1 | 1 | 3 |
| 13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 5 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 2 |
| 14 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 3 | 2 | 1 | 4 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 |
| 15 | 0 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 16 | 0 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| 17 | 2 | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 1 | 1 | 4 | 0 | 1 | 2 | 3 | 4 | 0 | 1 | 0 |
| 18 | 0 | 1 | 1 | 4 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| 19 | 2 | 1 | 1 | 5 | 1 | 1 | 1 | 5 | 1 | 1 | 3 | 4 | 1 | 1 | 0 | 1 | 2 | 1 | 1 |
| 20 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 6 | 1 | 1 | 3 | 1 | 2 | 2 | 4 | 0 | 1 | 1 | 1 |
| 21 | 2 | 1 | 3 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 6 | 5 | 1 | 1 | 1 |
| 22 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 5 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 2 | 0 | 1 | 0 |
| 23 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | 1 | 3 | 1 | 1 | 2 | 7 | 1 | 1 | 1 | 0 |
| 24 | 0 | 1 | 1 | 0 | 0 | 1 | 3 | 2 | 4 | 1 | 3 | 1 | 0 | 2 | 5 | 1 | 2 | 1 | 1 |
| 25 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 4 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| 26 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 1 | 1 | 3 | 0 | 0 | 3 | 2 | 1 | 0 |
| 27 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 2 | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 |
| 28 | 1 | 4 | 1 | 0 | 3 | 1 | 0 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 0 | 1 | 2 |
| 29 | 1 | 1 | 1 | 0 | 0 | 1 | 4 | 4 | 1 | 1 | 3 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 0 |
| 30 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | 0 |
| 31 | 1 | 4 | 1 | 0 | 0 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
| 32 | 1 | 1 | 5 | 0 | 0 | 1 | 3 | 4 | 6 | 1 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 |
| Marker | GenBank Accession | Primer sequence (5′–3′)(Forward) | Primer sequence (5′–3′)(Reverse) |
|---|---|---|---|
| ZJULM 1 | MF677780 | AAATTGGGTATCCATCTC | CATAAAACTTCCGTGAAA |
| ZJULM 2 | MF677781 | TATTTGGTCCAACAATAG | TTGAAAGTTCAATAAACC |
| ZJULM 4 | MF677782 | CTTTGGGCTTCTTCACTA | TTTGGGTGAAAGTTTTGT |
| ZJULM 5 | MF677783 | TCAACACTCTGCCAATTG | AGCCCCATGAACATAAAA |
| ZJULM 6 | MF677784 | TAAAAGTTCATTCATTACAC | GACATAACAAAATAGGATAA |
| ZJULM 7 | MF677785 | ATCTAAAATAAATTAACGGG | CAAATTTGGTTGAATTTACA |
| ZJULM 8 | MF677786 | ATTTGATTCGATGCTACC | GAGCTCTTCGGAATTTTA |
| ZJULM 9 | MF677787 | AAAGAAGCATAATCCCTT | TAACCTGCAATTCAATGT |
| ZJULM 10 | MF677788 | ATGGGAGTTGGGCTATTT | ATCAGCAGCAGTGTTTGG |
| ZJULM 11 | MF677789 | TCTTCCTCCCTCTTATCC | TCTGAATGGGGTTGGTTT |
| ZJULM 12 | MF677790 | GTGTTTGGTTATGAATTT | CCTTATAATTTCAATTCC |
| ZJULM 13 | MF677791 | TCTCCCTCCCTCTTGCTC | TGTAAACTTAACCCAAACCTC |
| ZJULM 14 | MF677792 | TTCTTCTCAGGCACTCCA | CACAAAGTACCAAGGTGG |
| ZJULM 15 | MF677793 | TGTTTGGATCTAAAGAAA | TAAACAACATGGATGAAT |
| ZJULM 16 | MF677794 | CTCTATGAGGTCTGTGGGAGA | GAGCTAAGCCCCAAAATC |
| ZJULM 17 | MF677795 | TTGTCTTTACTATTGGGA | ATTATCAAACATCCACAA |
| ZJULM 18 | MF677796 | TCAGGATTGTTAAGCCAGTT | CAATGACCAGCAATGACC |
| ZJULM 19 | MF677797 | GCACCTAAGCCAACCAAC | GGACAATGCATGTCACGA |
| ZJULM 22 | MF677798 | GAGAAGAAGACTCTGGGG | AAGAAAAGTGAAATCCCA |
| ZJULM 23 | MF677799 | GAAAAGTCGTTGACAACA | CAATTTCGTTTGAATGTT |
| ZJULM 24 | MF677800 | CGAATGTTAAAGAAACTT | CGAATGTTAAAGAAACTT |
| ZJULM 25 | MF677801 | AAAAGTCGTTGACAACAT | TACAATTTCGTTTGAATG |
| ZJULM 26 | MF677802 | AAACAGTTTCCCTTACCA | AATATCGTGGAGGTTGTC |
| ZJULM 27 | MF677803 | GGACACCAAAGTAAACATGC | CTAGTTTCATCAATTCCAAG |
| ZJULM 28 | MF677804 | ACTTGCTTATCAGAGTGGCA | ATGTTTGTCGGTAATGTTCG |
| ZJULM 29 | MF677805 | CCACCTGTAATGTTATCCAT | ATTTTGGTACGTTATCTGCT |
| ZJULM 30 | MF677806 | AGCAACTAAAATGAGGTAAA | TATTGATGGCATCCATCCTG |
| ZJULM 31 | MF677807 | GCCAACTCATAACAAGAATC | TAATCACCAACACCTTATTC |
| ZJULM 32 | MF677808 | GAAATGTGAAATCCCACG | TGGACGGAGTAGAGGTGA |
| ZJULM 33 | MF677809 | TAGCCGTTCGTTTTCATT | CACCGACATTCTAAATCCTG |
| ZJULM 34 | MF677810 | CATGGCGGCTATGAAGGC | TCCGCACAGTGACAGAGTGGT |
| ZJULM 35 | MF677811 | TTATGTCTGTCCCGTTCA | ATACCTTATCTTTGTGCC |
| ZJULM 38 | MF677812 | GGGGAGAAATAAGAAATAG | TTCGCTTCGTGGTGTTGG |
| ZJULM 40 | MF677813 | TATCCAATAAGCTTGAAG | AAACTATCGCATGTAATG |
| ZJULM 43 | MF677814 | CTACCCGTGAGAATTTGA | CACTACTTCCACCCACAA |
| ZJULM 48 | MF677815 | TAGAAAGGAAAGGAGGAA | TTCAAGAGTTCAGGGTTT |
| ZJULM 49 | MF677816 | GAGAAAGATAATTGAAAGGGAT | GTGCTGCCATACGGTTAG |
| ZJULM 52 | MF677817 | ATCTAAAATTTAAAGGGG | TAGACCATAATACCCCTT |
| ZJULM 54 | MF677818 | TGTTGTTATGAATCGGTGAA | TAGGCAAAGGAAAGTTGG |
| ZJULM 56 | MF677819 | TGGCGGCGGAGCAGTGAA | ACCACCCGTAGGGCGTGTCC |
| ZJULM 57 | MF677820 | TTCTTCTCCCTCTTTGCT | ACAGTCACCGCCTCATAT |
| ZJULM 58 | MF677821 | GTATCGTATCGGGTGCCT | TTCCTTTCCACATGCCTC |
| ZJULM 60 | MF677822 | ATTTCTGTTAATTTGGTTCC | CAATCGAATAAAAGGTCAA |
| ZJULM 62 | MF677823 | TTTTCAAAGTTCAAGGAC | TTAGTGTCACGTCAGCAT |
| ZJULM 63 | MF677824 | CAGGCGAAGCAAAGGATT | TGATGGTCTGACGGAGGC |
| ZJULM 64 | MF677825 | TTTGTCACAATCCCACCT | GAATACGCAGCCTTCTTT |
| ZJULM 65 | MF677826 | AGAATGATTTACCCGTAG | AGAGGAGGAACTTTTGAT |
| ZJULM 68 | MF677827 | CCCCTCCCCTCCAAAATA | TTGCCCAGGAACGAACTT |
| ZJULM 69 | MF677828 | TCATTCCTACCGAAAGTA | AACGGACCCTTATACTTG |
| ZJULM 70 | MF677829 | AAGCGGGAGCTAAGAATG | GCTGGAATGTTGGGAGAA |
| ZJULM 72 | MF677830 | ACACCGTAACAGATCAAA | CTCATTCTTTCCCTTTCT |
| ZJULM 74 | MF677831 | ATCTAAAATTTAAAGGGG | TAGACCATAATACCCCTT |
| ZJULM 75 | MF677832 | TGTTGTTATGAATCGGTGAA | TAGGCAAAGGAAAGTTGG |
| ZJULM 76 | MF677833 | AACCCACAGAATAAAGATG | GAAGAAGCTCCTACCTGA |
| ZJULM 80 | MF677834 | TCAATGCCAGTGTCTCAA | GCTTCTTATTGGACCTATTT |
| No. | Accession | Region | No. | Accession | Region |
|---|---|---|---|---|---|
| 1 | Zheda 1 | Hangzhou, Zhejiang | 17 | Zheda 7 | Hangzhou, Zhejiang |
| 2 | Zheda 2 | Hangzhou, Zhejiang | 18 | Zheda 8 | Hangzhou, Zhejiang |
| 3 | Zheda 23 | Hangzhou, Zhejiang | 19 | Zheda h101 | Hangzhou, Zhejiang |
| 4 | Zheda 56 | Hangzhou, Zhejiang | 20 | Zheda h100 | Hangzhou, Zhejiang |
| 5 | Zheda 10 | Hangzhou, Zhejiang | 21 | Zheda 57N | Hangzhou, Zhejiang |
| 6 | Zheda 81 | Hangzhou, Zhejiang | 22 | Zheda 72 | Hangzhou, Zhejiang |
| 7 | Zheda 83 | Hangzhou, Zhejiang | 23 | Zheda 22 | Hangzhou, Zhejiang |
| 8 | Zheda 84 | Hangzhou, Zhejiang | 24 | Zheda 70 | Hangzhou, Zhejiang |
| 9 | Shuangjiannaihan1 | Heshan, Guangdong | 25 | Zheda 5 | Hangzhou, Zhejiang |
| 10 | Jizaoxiangyu | Changsha, Hunan | 26 | Zheda h6 | Hangzhou, Zhejiang |
| 11 | Jinxiuzaojia | Lanzhou, Gansu | 27 | Zheda h7 | Hangzhou, Zhejiang |
| 12 | Sanbier | Changde, Hunan | 28 | Zheda 85 | Hangzhou, Zhejiang |
| 13 | Baimeichunxiang | Changsha, Hunan | 29 | Zheda 86 | Hangzhou, Zhejiang |
| 14 | Jipinduanbang | Changsha, Hunan | 30 | Zheda 87 | Hangzhou, Zhejiang |
| 15 | Lvbaoshi | Lanzhou, Gansu | 31 | Zheda 88 | Hangzhou, Zhejiang |
| 16 | Chunjianlv | Lanzhou, Gansu | 32 | Zheda h4 | Hangzhou, Zhejiang |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.; Yin, M.; Zhang, Q.; Gong, D.; Jia, X.; Guan, Y.; Hu, J. Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes. Int. J. Mol. Sci. 2017, 18, 1942. https://doi.org/10.3390/ijms18091942
An J, Yin M, Zhang Q, Gong D, Jia X, Guan Y, Hu J. Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes. International Journal of Molecular Sciences. 2017; 18(9):1942. https://doi.org/10.3390/ijms18091942
Chicago/Turabian StyleAn, Jianyu, Mengqi Yin, Qin Zhang, Dongting Gong, Xiaowen Jia, Yajing Guan, and Jin Hu. 2017. "Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes" International Journal of Molecular Sciences 18, no. 9: 1942. https://doi.org/10.3390/ijms18091942