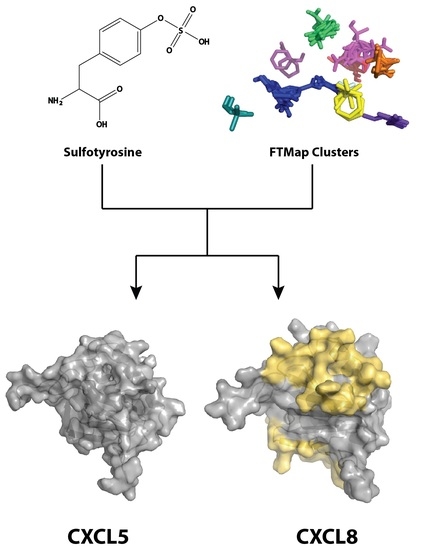
Differences in Sulfotyrosine Binding amongst CXCR1 and CXCR2 Chemokine Ligands
Abstract
:1. Introduction
2. Results
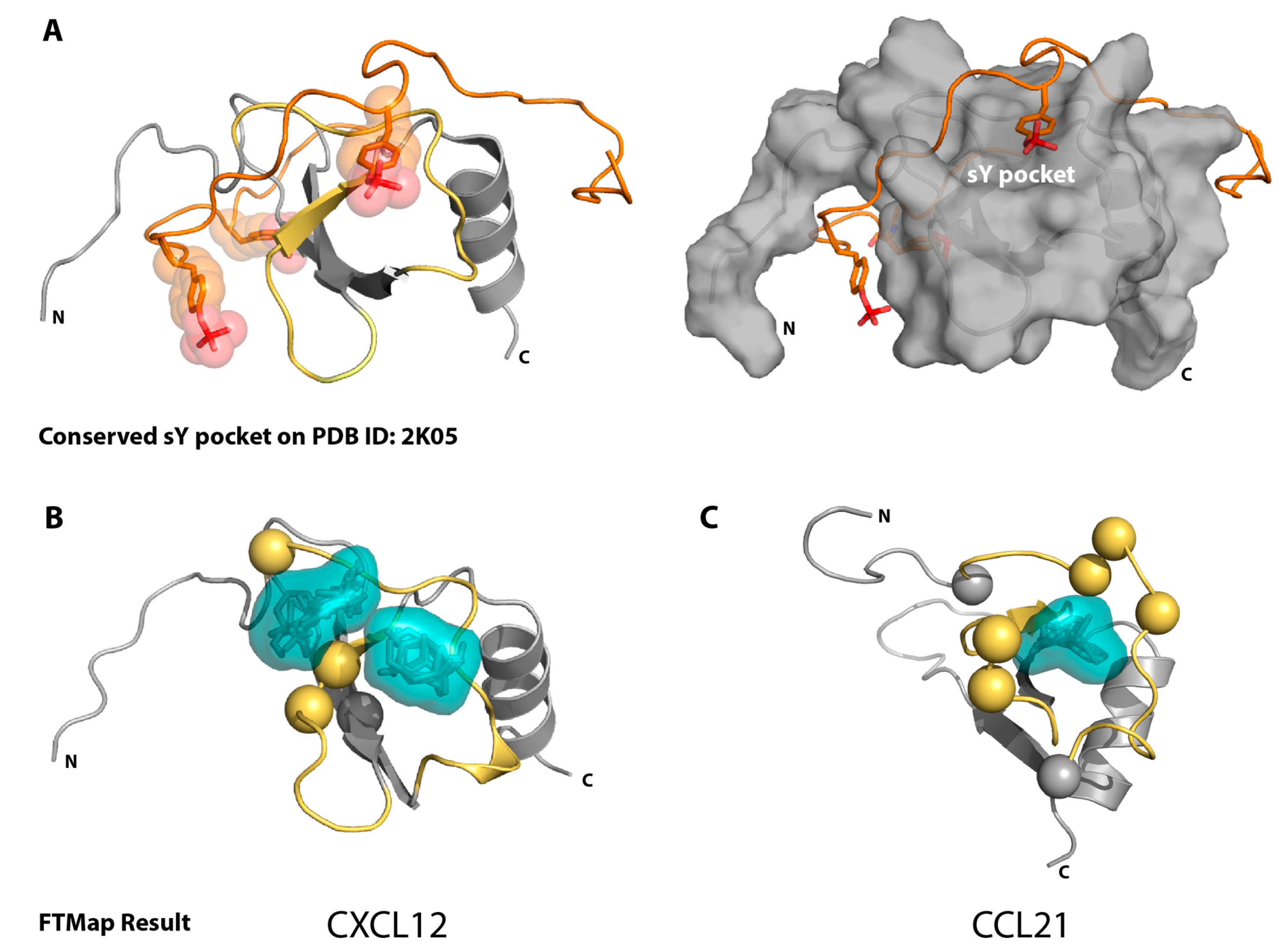
2.1. Sulfotyrosine Recognition Sites Correspond to Predicted Chemokine Hot Spots
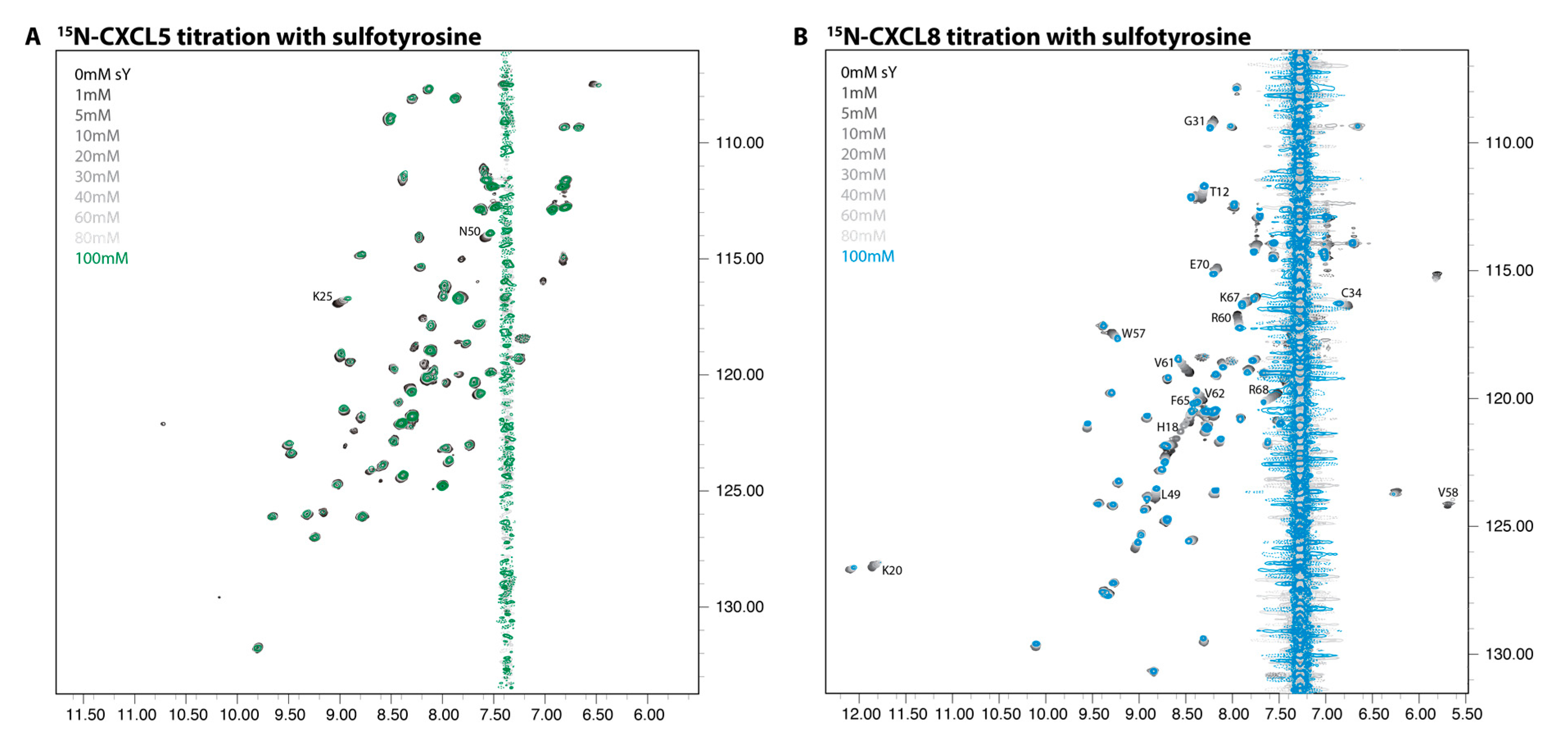
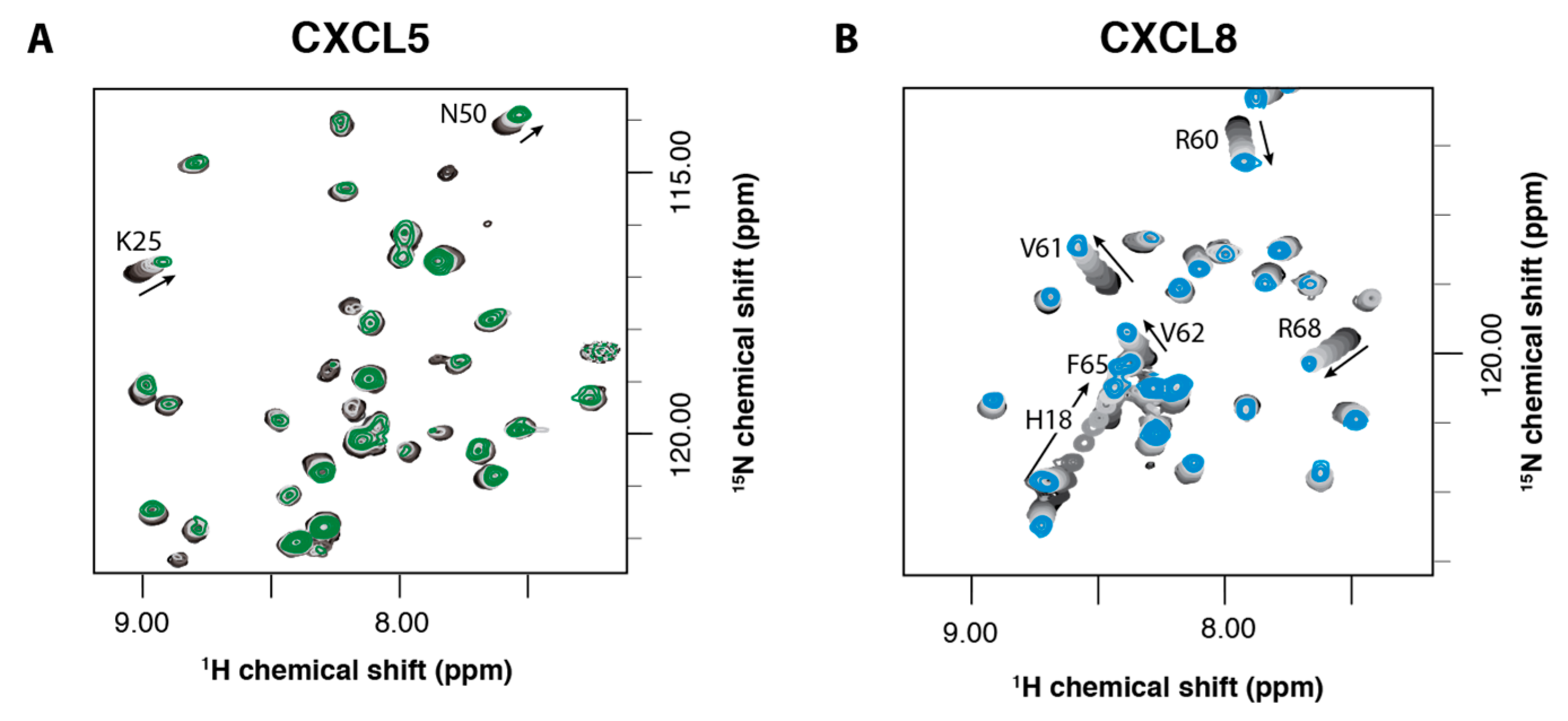
2.2. NMR Titration Studies with Sulfotyrosine
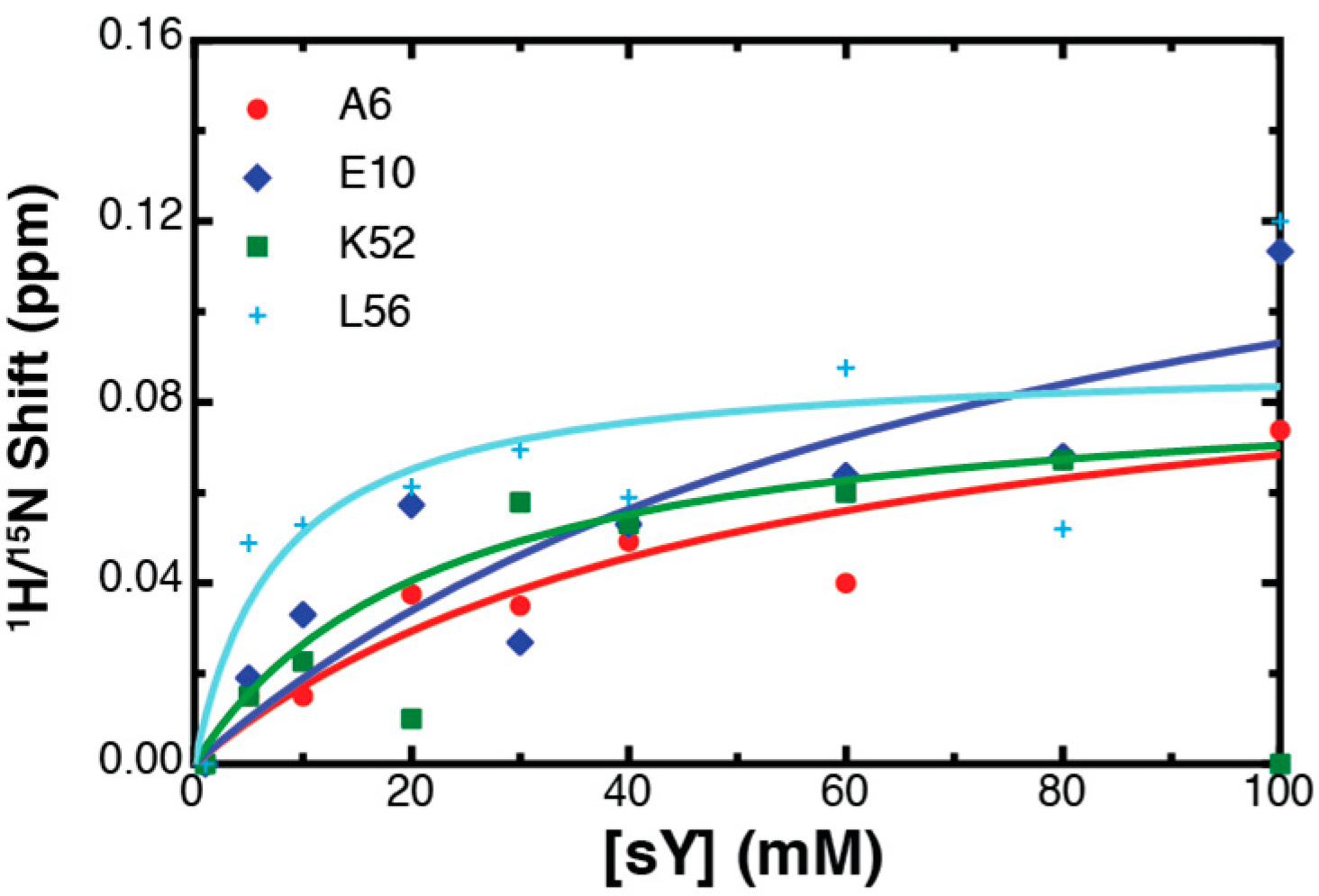
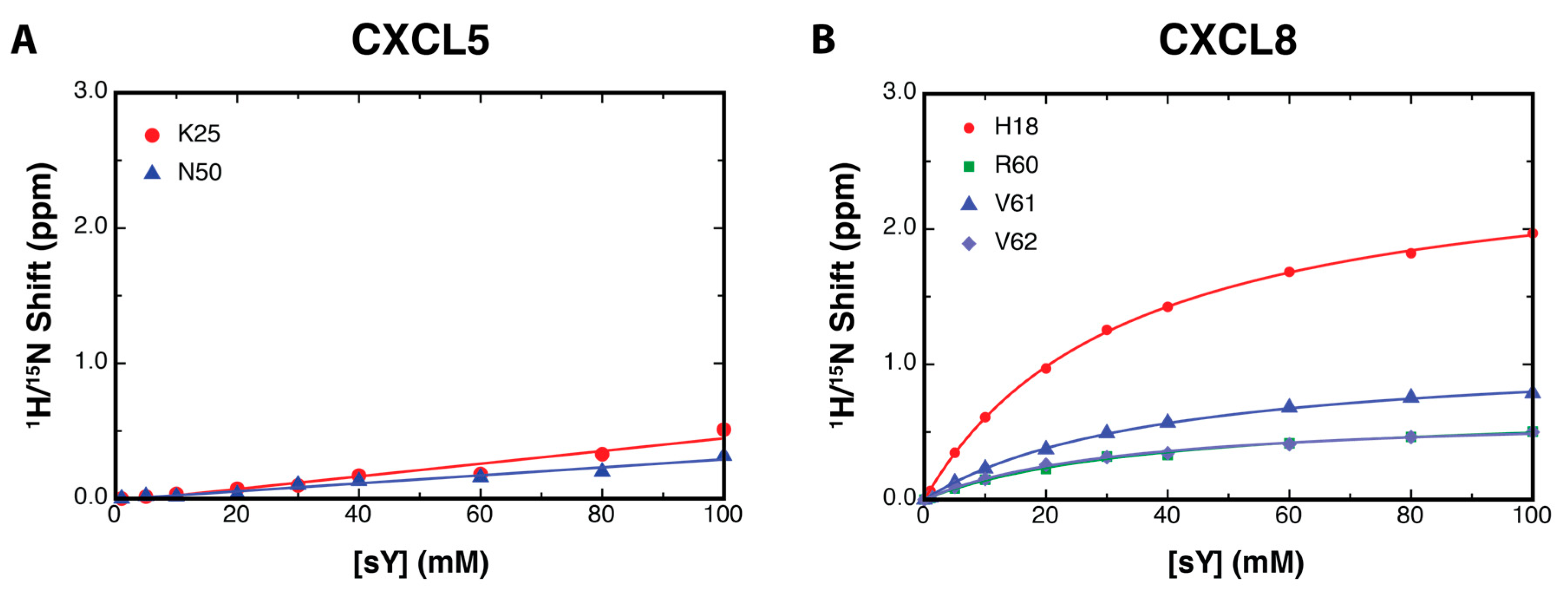
2.3. Binding Affinity to Sulfotyrosine
3. Discussion
4. Materials and Methods
4.1. FTMap
4.2. Protein Expression and Purification
4.3. NMR Spectroscopy
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CXCL | CXC ligand |
| CXCR | CXC receptor |
| CCL | CC ligand |
| GPCR | G-protein Coupled Receptor |
| sY | Sulfotyrosine |
| NMR | Nuclear Magnetic Resonance |
| HSQC | Heteronuclear Single Quantum Coherence |
| CSP | Chemical Shift Perturbation |
| ELR+ | Glutamate-leucine-arginine positive |
| GAG | Glycosaminoglycan |
| SPR | Surface Plasmon Resonance |
Appendix A

References
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O.; Nomiyama, H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Salanga, C.L.; Handel, T.M. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol. Cell Biol. 2015, 93, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Chuang, S.; Hou, X.; Shoham, M.; Zhu, J.Z. Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. New Biotechnology 2009, 25, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Ludeman, J.P.; Stone, M.J. The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 2014, 171, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.; Veldkamp, C.T.; Peterson, F.C.; Chait, B.T.; Volkman, B.F.; Sakmar, T.P. Sequential tyrosine sulfation of cxcr4 by tyrosylprotein sulfotransferases. Biochemistry 2008, 47, 11251–11262. [Google Scholar] [CrossRef] [PubMed]
- Jen, C.H.; Leary, J.A. A competitive binding study of chemokine, sulfated receptor, and glycosaminoglycan interactions by nano-electrospray ionization mass spectrometry. Anal. Biochem. 2010, 407, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.S.; Zhu, J.Z.; Widlanski, T.S.; Stone, M.J. Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem. Biol. 2009, 16, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Ludeman, J.P.; Wedderburn, J.; Canals, M.; Hall, P.; Butler, S.J.; Taleski, D.; Christopoulos, A.; Hickey, M.J.; Payne, R.J.; et al. Tyrosine sulfation of chemokine receptor ccr2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (mcp-1). J. Biol. Chem. 2013, 288, 10024–10034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Z.; Millard, C.J.; Ludeman, J.P.; Simpson, L.S.; Clayton, D.J.; Payne, R.J.; Widlanski, T.S.; Stone, M.J. Tyrosine sulfation influences the chemokine binding selectivity of peptides derived from chemokine receptor ccr3. Biochemistry 2011, 50, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; De la Cruz, N.B.; Haugner, J.C., 3rd; Basnet, H.; Sakmar, T.P.; Volkman, B.F. Structural basis of cxcr4 sulfotyrosine recognition by the chemokine sdf-1/cxcl12. Sci. Signal. 2008, 1. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; Sakmar, T.P.; Volkman, B.F. Recognition of a cxcr4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (sdf-1alpha/cxcl12). J. Mol. Biol. 2006, 359, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Duma, L.; Haussinger, D.; Rogowski, M.; Lusso, P.; Grzesiek, S. Recognition of rantes by extracellular parts of the ccr5 receptor. J. Mol. Biol. 2007, 365, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Getschman, A.E.; Butler, S.J.; Taleski, D.; Stephens, B.; Kufareva, I.; Handel, T.M.; Payne, R.J.; Volkman, B.F. Sulfopeptide probes of the cxcr4/cxcl12 interface reveal oligomer-specific contacts and chemokine allostery. ACS Chem. Biol. 2013, 8, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Heroux, M.S.; Veldkamp, C.T.; Peterson, F.C.; Volkman, B.F. Sulfotyrosine recognition as marker for druggable sites in the extracellular space. Int. J. Mol. Sci. 2011, 12, 3740–3756. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.J.; Ludeman, J.P.; Canals, M.; Bridgford, J.L.; Hinds, M.G.; Clayton, D.J.; Christopoulos, A.; Payne, R.J.; Stone, M.J. Structural basis of receptor sulfotyrosine recognition by a CC chemokine: The N-terminal region of CCR3 bound to CCL11/Eotaxin-1. Structure 2014, 22, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Stillie, R.; Farooq, S.M.; Gordon, J.R.; Stadnyk, A.W. The functional significance behind expressing two IL-8 receptor types on pmn. J. Leukoc. Biol. 2009, 86, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Baggiolini, M.; Dewald, B.; Moser, B. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv. Immunol. 1994, 55, 97–179. [Google Scholar] [PubMed]
- Dwinell, M.B.; Kagnoff, M.F. Mucosal immunity. Curr. Opin. Gastroenterol. 1999, 15, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Kagnoff, M.F.; Eckmann, L. Epithelial cells as sensors for microbial infection. J. Clin. Invest. 1997, 100, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Delgado, M.B.; Jones, S.A.; Dewald, B.; Clark-Lewis, I.; Baggiolini, M. Granulocyte chemotactic protein 2 acts via both il-8 receptors, cxcr1 and cxcr2. Eur. J. Immunol. 1998, 28, 164–170. [Google Scholar] [CrossRef]
- Koch, A.E.; Kunkel, S.L.; Harlow, L.A.; Mazarakis, D.D.; Haines, G.K.; Burdick, M.D.; Pope, R.M.; Walz, A.; Strieter, R.M. Epithelial neutrophil activating peptide-78: A novel chemotactic cytokine for neutrophils in arthritis. J. Clin. Invest. 1994, 94, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Chavey, C.; Lazennec, G.; Lagarrigue, S.; Clape, C.; Iankova, I.; Teyssier, J.; Annicotte, J.S.; Schmidt, J.; Mataki, C.; Yamamoto, H.; et al. Cxc ligand 5 is an adipose-tissue derived factor that links obesity to insulin resistance. Cell. Metab. 2009, 9, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Dawes, J.M.; Calvo, M.; Perkins, J.R.; Paterson, K.J.; Kiesewetter, H.; Hobbs, C.; Kaan, T.K.; Orengo, C.; Bennett, D.L.; McMahon, S.B. Cxcl5 mediates uvb irradiation-induced pain. Sci Transl. Med. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.K.; Murphy, P.M. The CXC chemokines growth-regulated oncogene (gro) alpha, grobeta, grogamma, neutrophil-activating peptide-2, and epithelial cell-derived neutrophil-activating peptide-78 are potent agonists for the type b, but not the type a, human interleukin-8 receptor. J. Biol. Chem. 1996, 271, 20545–20550. [Google Scholar] [CrossRef] [PubMed]
- Monigatti, F.; Gasteiger, E.; Bairoch, A.; Jung, E. The sulfinator: Predicting tyrosine sulfation sites in protein sequences. Bioinformatics 2002, 18, 769–770. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Lee, T.Y.; Shien, D.M.; Hsu, J.B.; Horng, J.T.; Hsu, P.C.; Wang, T.Y.; Huang, H.D.; Pan, R.L. Incorporating support vector machine for identifying protein tyrosine sulfation sites. J. Comput. Chem. 2009, 30, 2526–2537. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Louie, S.; Hsu, W.; Yu, K.M.; Nicholas, H.B., Jr.; Rosenquist, G.L. Tyrosine sulfation is prevalent in human chemokine receptors important in lung disease. Am. J. Respir. Cell. Mol. Biol. 2008, 38, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.W.; Lewandowski, E.M.; Moussouras, N.A.; Kroeck, K.G.; Volkman, B.F.; Veldkamp, C.T.; Chen, Y. Crystallographic structure of truncated CCL21 and the putative sulfotyrosine-binding site. Biochemistry 2016, 55, 5746–5753. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.W.; Nevins, A.M.; Qiao, Z.; Liu, Y.; Getschman, A.E.; Vankayala, S.L.; Kemp, M.T.; Peterson, F.C.; Li, R.; Volkman, B.F.; et al. Structure-based identification of novel ligands targeting multiple sites within a chemokine-g-protein-coupled-receptor interface. J. Med. Chem. 2016, 59, 4342–4351. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Grove, L.E.; Hall, D.R.; Bohnuud, T.; Mottarella, S.E.; Luo, L.; Xia, B.; Beglov, D.; Vajda, S. The ftmap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015, 10, 733–755. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Chuang, G.Y.; Cencic, R.; Brenke, R.; Grove, L.E.; Beglov, D.; Pelletier, J.; Whitty, A.; Vajda, S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc. Natl. Acad. Sci. USA 2011, 108, 13528–13533. [Google Scholar] [CrossRef] [PubMed]
- Skelton, N.J.; Quan, C.; Reilly, D.; Lowman, H. Structure of a cxc chemokine-receptor fragment in complex with interleukin-8. Structure 1999, 7, 157–168. [Google Scholar] [CrossRef]
- Joseph, P.R.; Rajarathnam, K. Solution nmr characterization of wt cxcl8 monomer and dimer binding to cxcr1 n-terminal domain. Protein Sci. 2015, 24, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.K.; Eckmann, L.; Panja, A.; Kagnoff, M.F. Differential and regulated expression of c-x-c, c-c, and c-chemokines by human colon epithelial cells. Gastroenterology 1997, 113, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Johanesen, P.A.; Dwinell, M.B. Flagellin-independent regulation of chemokine host defense in campylobacter jejuni-infected intestinal epithelium. Infect. Immun. 2006, 74, 3437–3447. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, E.L.; Karavitis, J.; Deburghgraeve, C.; Ramirez, L.; Kovacs, E.J. Prolonged chemokine expression and excessive neutrophil infiltration in the lungs of burn-injured mice exposed to ethanol and pulmonary infection. Shock 2011, 35, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Proost, P.; De Wolf-Peeters, C.; Conings, R.; Opdenakker, G.; Billiau, A.; Van Damme, J. Identification of a novel granulocyte chemotactic protein (gcp-2) from human tumor cells. In vitro and in vivo comparison with natural forms of gro, IP-10, and IL-8. J. Immunol. 1993, 150, 1000–1010. [Google Scholar] [PubMed]
- Kunkel, S.L.; Lukacs, N.; Strieter, R.M. Expression and biology of neutrophil and endothelial cell-derived chemokines. Semin. Cell. Biol. 1995, 6, 327–336. [Google Scholar] [CrossRef]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Bannert, N.; Craig, S.; Farzan, M.; Sogah, D.; Santo, N.V.; Choe, H.; Sodroski, J. Sialylated o-glycans and sulfated tyrosines in the NH2-terminal domain of cc chemokine receptor 5 contribute to high affinity binding of chemokines. J. Exp. Med. 2001, 194, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.R.; Mosier, P.D.; Desai, U.R.; Rajarathnam, K. Solution nmr characterization of chemokine cxcl8/il-8 monomer and dimer binding to glycosaminoglycans: Structural plasticity mediates differential binding interactions. Biochem. J. 2015, 472, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Sepuru, K.M.; Nagarajan, B.; Desai, U.R.; Rajarathnam, K. Molecular basis of chemokine CXCL5-glycosaminoglycan interactions. J. Biol. Chem. 2016, 291, 20539–20550. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Sepuru, K.M.; Rajarathnam, K. Structural basis of native CXCL7 monomer binding to CXCR2 receptor n-domain and glycosaminoglycan heparin. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Sepuru, K.M.; Rajarathnam, K. CXCL1/MGSA is a novel glycosaminoglycan (gag)-binding chemokine: Structural evidence for two distinct non-overlapping binding domains. J. Biol. Chem. 2016, 291, 4247–4255. [Google Scholar] [CrossRef] [PubMed]
- Wuyts, A.; Proost, P.; Lenaerts, J.P.; Ben-Baruch, A.; Van Damme, J.; Wang, J.M. Differential usage of the cxc chemokine receptors 1 and 2 by interleukin-8, granulocyte chemotactic protein-2 and epithelial-cell-derived neutrophil attractant-78. Eur. J. Biochem. 1998, 255, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Sepuru, K.M.; Poluri, K.M.; Rajarathnam, K. Solution structure of cxcl5--a novel chemokine and adipokine implicated in inflammation and obesity. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.J.; Liou, J.W.; Chang, C.C.; Chung, Y.; Lin, L.F.; Hsu, H.J. Peptides derived from cxcl8 based on in silico analysis inhibit cxcl8 interactions with its receptor cxcr1. Sci. Rep. 2015, 5, 18638. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger, LLC. The pymol molecular graphics system. version 1.7.0.3. 2010. [Google Scholar]
- Veldkamp, C.T.; Koplinski, C.A.; Jensen, D.R.; Peterson, F.C.; Smits, K.M.; Smith, B.L.; Johnson, S.K.; Lettieri, C.; Buchholz, W.G.; Solheim, J.C.; et al. Production of recombinant chemokines and validation of refolding. Methods Enzymol. 2016, 570, 539–565. [Google Scholar] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe: A multidimensional spectral processing system based on unix pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, B.L.; Gronenborn, A.M.; Clore, G.M. Analysis of the backbone dynamics of interleukin-8 by 15n relaxation measurements. J. Mol. Biol. 1993, 230, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.L.J. (Ed.) The Computer Aided Resonance Assignment/Tutorial, 1st ed.; CANTINA Verlag: Goldau, Switzerland, 2004. [Google Scholar]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moussouras, N.A.; Getschman, A.E.; Lackner, E.R.; Veldkamp, C.T.; Dwinell, M.B.; Volkman, B.F. Differences in Sulfotyrosine Binding amongst CXCR1 and CXCR2 Chemokine Ligands. Int. J. Mol. Sci. 2017, 18, 1894. https://doi.org/10.3390/ijms18091894
Moussouras NA, Getschman AE, Lackner ER, Veldkamp CT, Dwinell MB, Volkman BF. Differences in Sulfotyrosine Binding amongst CXCR1 and CXCR2 Chemokine Ligands. International Journal of Molecular Sciences. 2017; 18(9):1894. https://doi.org/10.3390/ijms18091894
Chicago/Turabian StyleMoussouras, Natasha A., Anthony E. Getschman, Emily R. Lackner, Christopher T. Veldkamp, Michael B. Dwinell, and Brian F. Volkman. 2017. "Differences in Sulfotyrosine Binding amongst CXCR1 and CXCR2 Chemokine Ligands" International Journal of Molecular Sciences 18, no. 9: 1894. https://doi.org/10.3390/ijms18091894