Systematic Selection of Reference Genes for the Normalization of Circulating RNA Transcripts in Pregnant Women Based on RNA-Seq Data
Abstract
:1. Introduction
2. Results
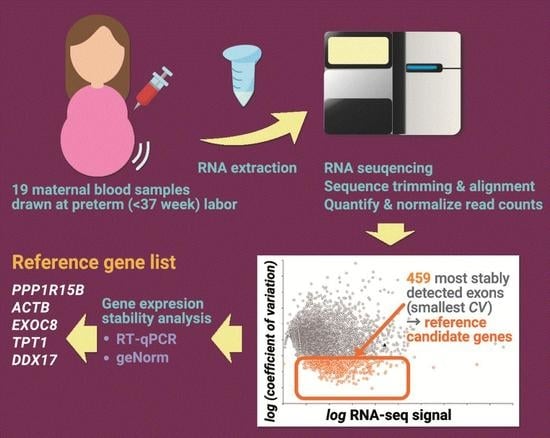
2.1. Whole-Transcriptome Profiling of Maternal Blood Samples
2.1.1. Overall Results of the RNA-seq Experiment
2.1.2. Identification of Exons That Are Stably Detected in Maternal Blood
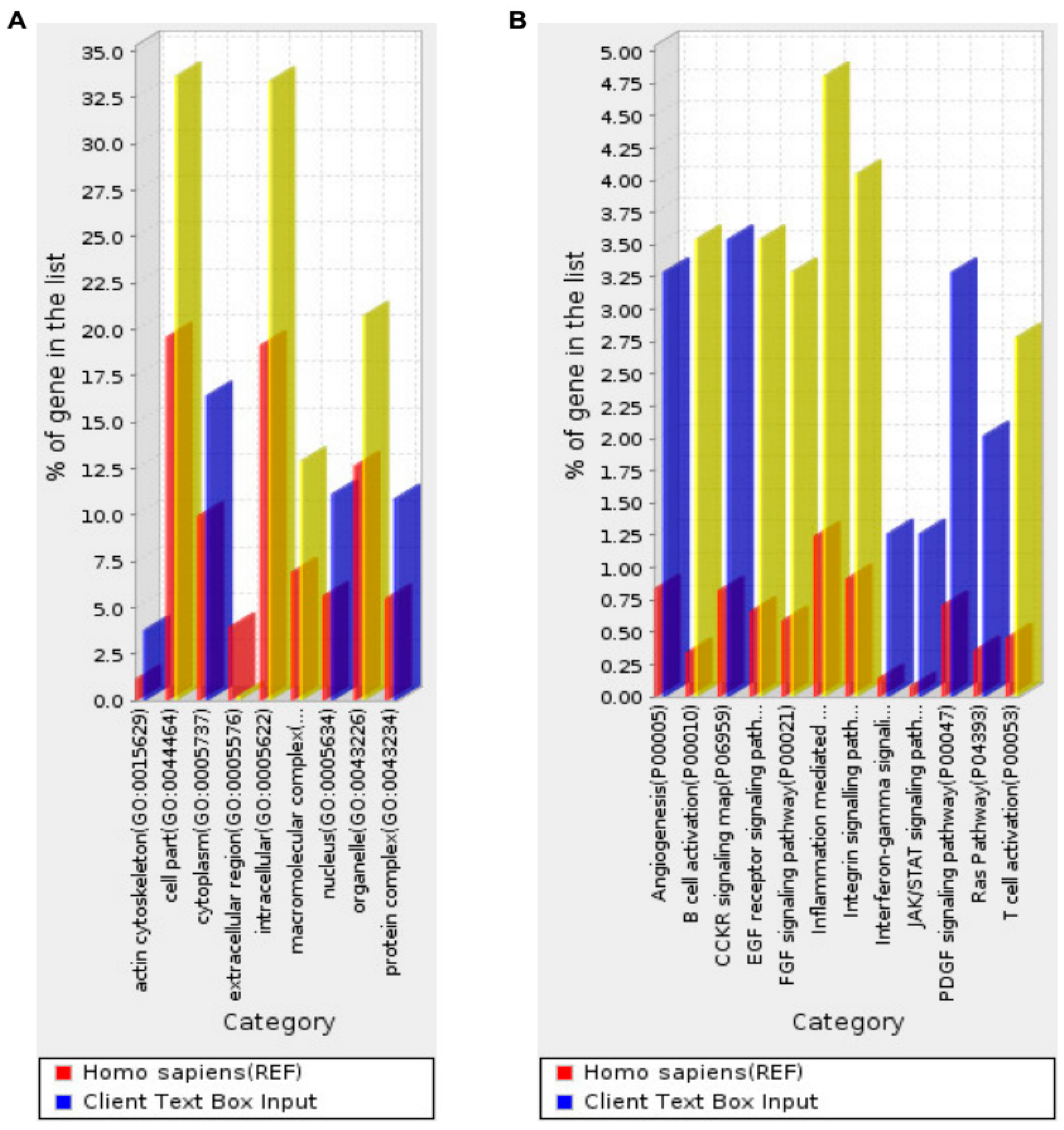
2.1.3. Pathways and Functional Annotation Terms Associated with the Genes That Are Stably Detected in Maternal Blood
2.1.4. Shortlisting of Reference Genes from the RNA-Sequencing Data and the Literature
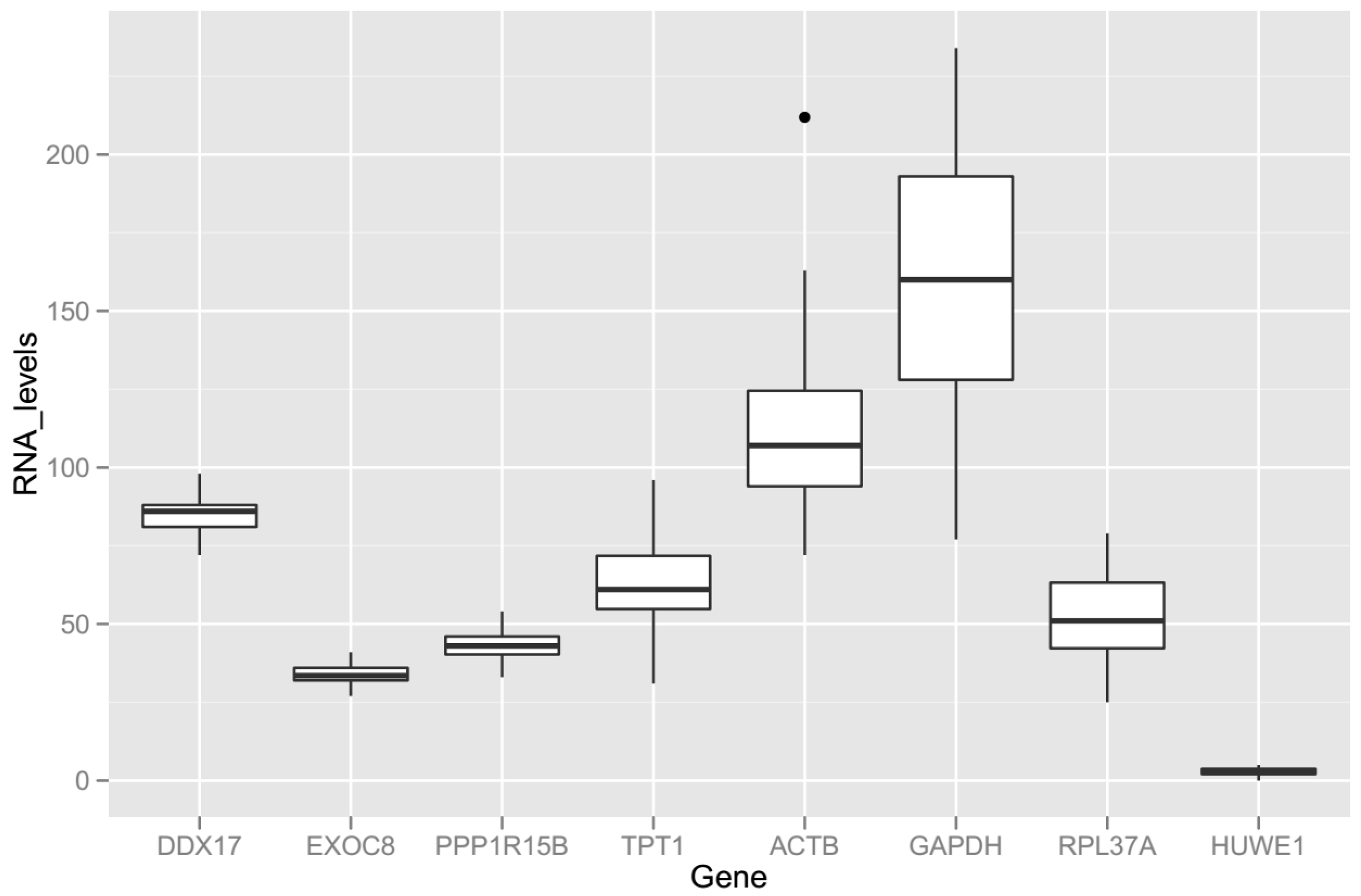
2.1.5. Exon-Level RNA-Sequencing Data in Selected Genes
2.2. Gene Expression Stability Analysis
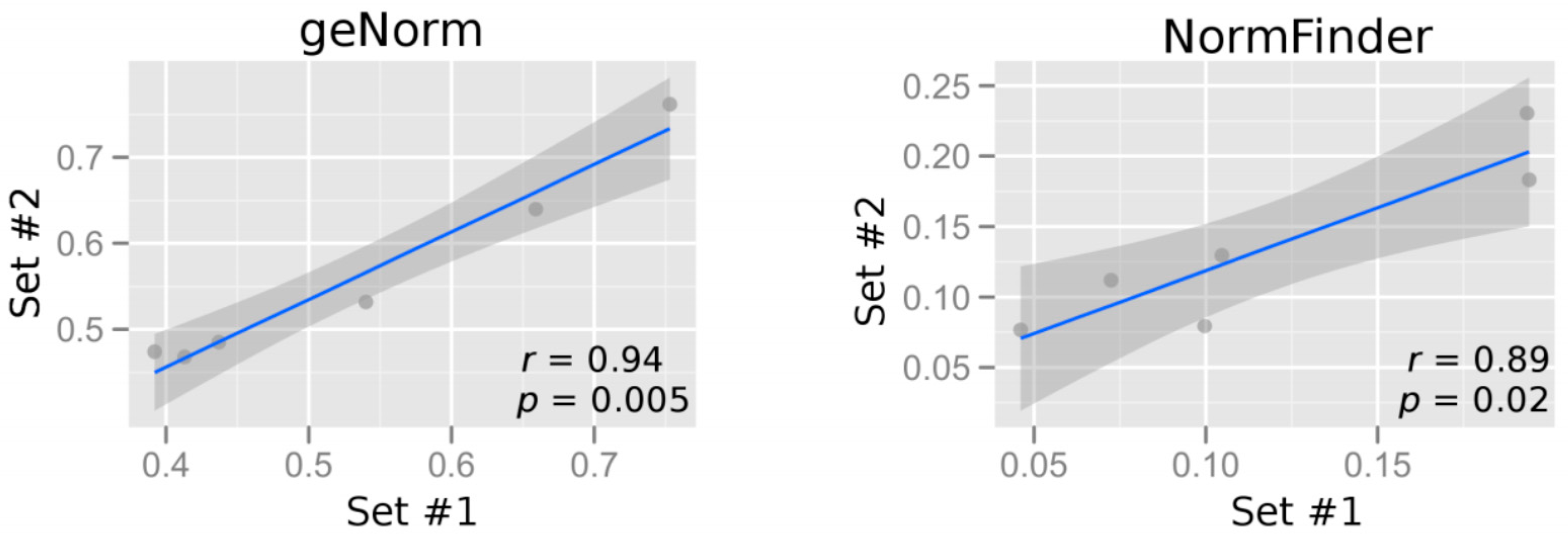
2.2.1. Comparing Complementary Algorithms in Analyzing Gene Expression Stability
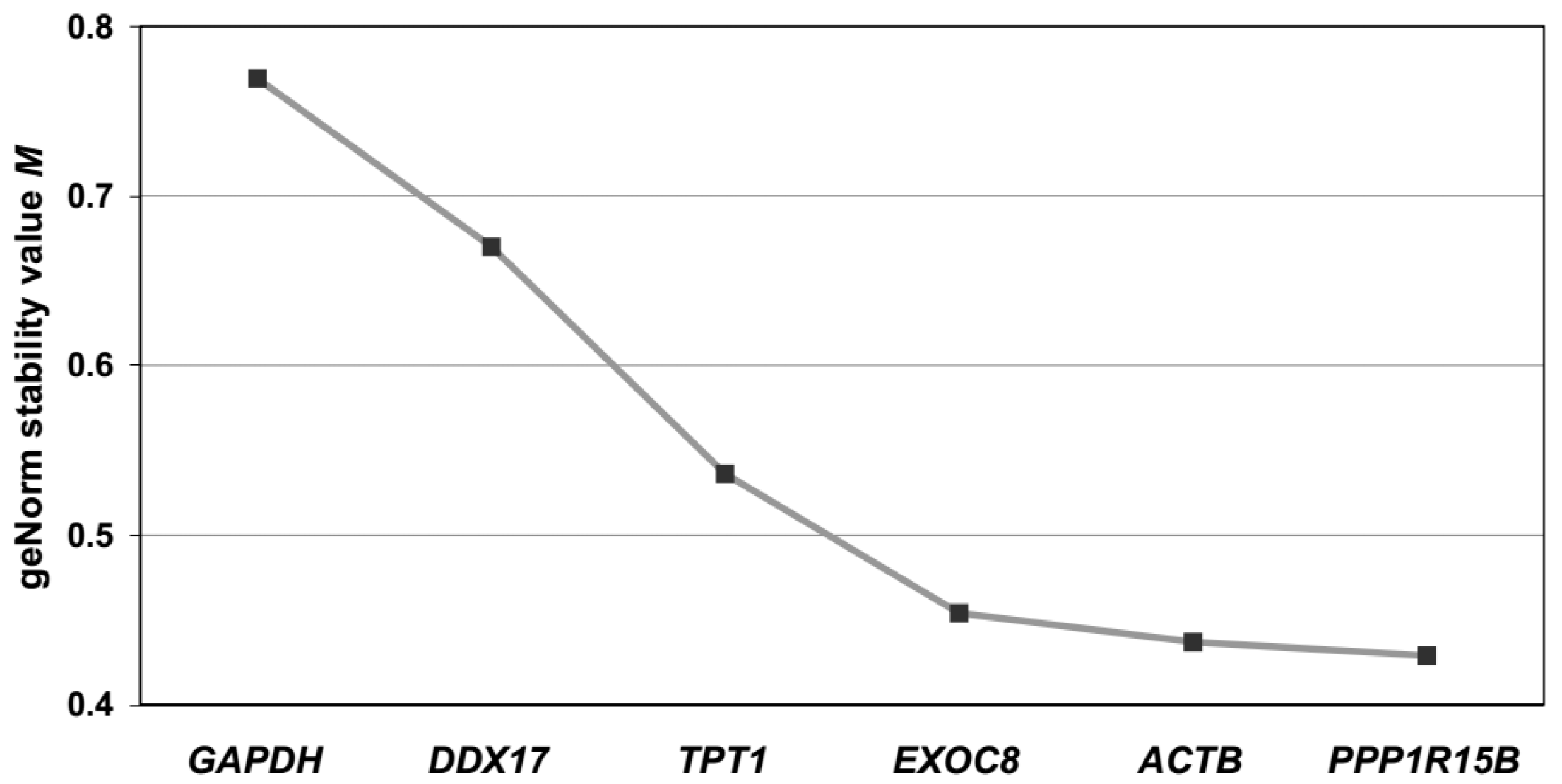
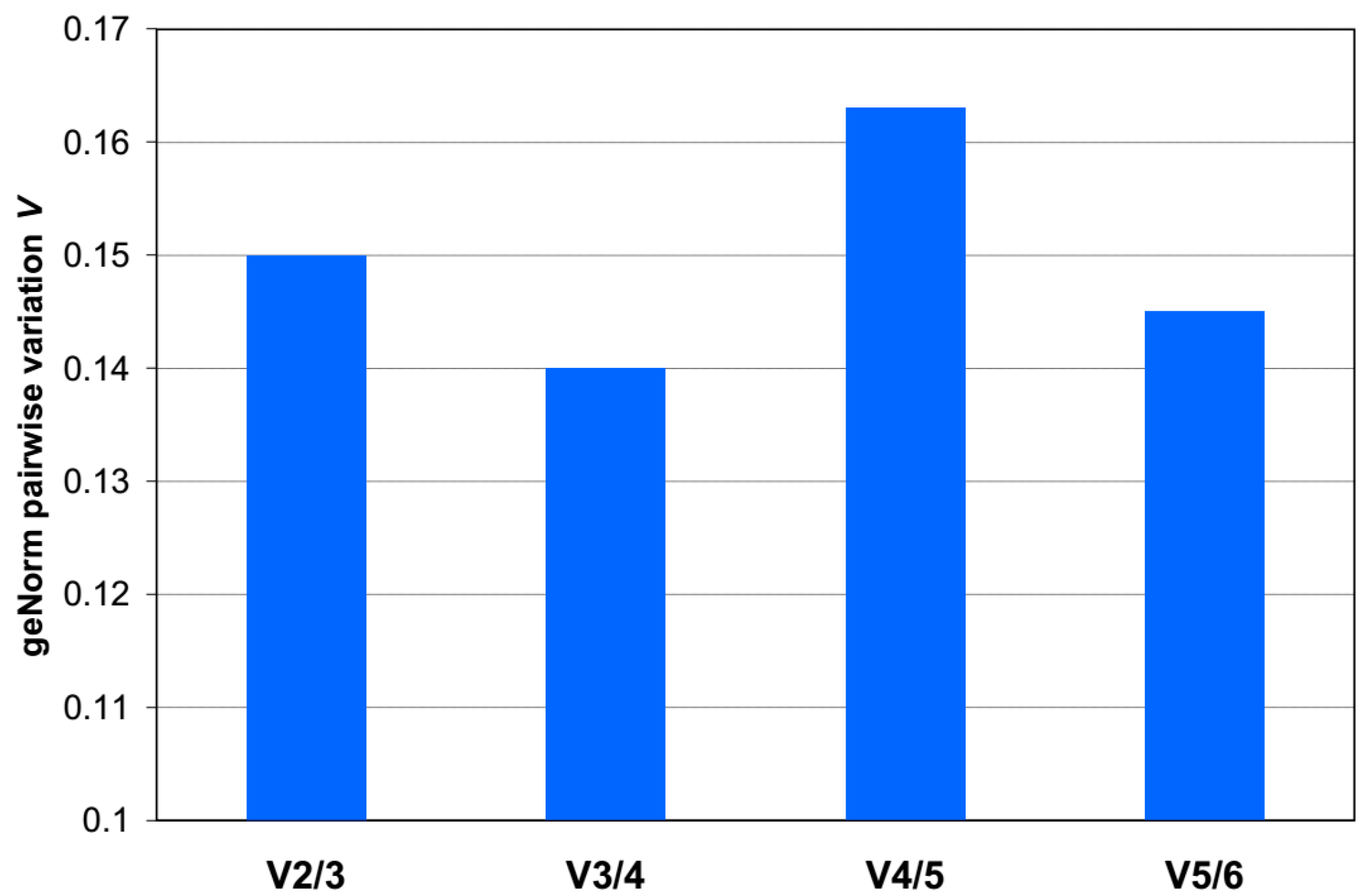
2.2.2. Assessing Gene Expression Stability in Maternal Blood Samples
3. Discussion
4. Materials and Methods
4.1. Recruitment of Study Subjects
4.2. Blood Collection and RNA Extraction
4.3. RNA-Sequencing
4.4. RT-qPCR
4.5. Other Data Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ssRNA-seq | Strand-specific RNA sequencing |
| RT | Reverse transcription |
| qPCR | Quantitative polymerse chain reaction |
| sPTB | Spontaneous preterm birth |
| TB | Term birth |
| CV | Coefficient of variation |
References
- Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology 1992, 10, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [PubMed]
- Romero, R.; Dey, S.K.; Fisher, S.J. Preterm labor: One syndrome, many causes. Science 2014, 345, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.C.; Lee, W.S.; Ting, Y.H.; Chan, O.K.; Lee, S.W.Y.; Leung, T.Y. Systematic identification of spontaneous preterm birth-associated RNA transcripts in maternal plasma. PLoS ONE 2012, 7, e34328. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.C.; Cheung, A.C.Y.; Cheung, K.W.C.; Leung, T.Y.; Lee, W.S. Biomarkers for Premature Birth. U.S. Patent Application Number 14/902,542, 29 December 2016. [Google Scholar]
- Tsuang, M.T.; Nossova, N.; Yager, T.; Tsuang, M.M.; Guo, S.C.; Shyu, K.G.; Glatt, S.J.; Liew, C.C. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: A preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005, 133, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.C.; Chang, C.W.; Chen, C.R.; Tsai, M.L.; Shu, W.Y.; Li, C.Y.; Hsu, I.C. Identification of reference genes across physiological states for qRT-PCR through microarray meta-analysis. PLoS ONE 2011, 6, e17347. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar] [PubMed]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S. The reactome pathway knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Cardenas, I.; Abrahams, V.; Guller, S. Inflammation and pregnancy: The role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 2011, 1221, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Itoh, N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005, 1. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.; Aplin, A.E.; Alahari, S.K.; Juliano, R.L. Integrin signaling and cell growth control. Curr. Opin. Cell Biol. 1998, 10, 220–231. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.-S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Watson, M. Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol. 2015, 16, 177. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.C.; Shing, T.K.F.; Hung, E.C.W.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.K.; Lo, Y.M.D. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Lui, Y.Y.N.; Chik, K.W.; Chiu, R.W.K.; Ho, C.Y.; Lam, C.W.K.; Lo, Y.M.D. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F. Proteogenomics connects somatic mutations to signaling in breast cancer. Nature 2016, 534, 55. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Nie, J.; Sicotte, H.; Li, Y.; Eckel-Passow, J.E.; Dasari, S.; Vedell, P.T.; Barman, P.; Wang, L.; Weinshiboum, R. Measure transcript integrity using RNA-seq data. BMC Bioinform. 2016, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155. [Google Scholar] [PubMed]
- Zhang, J.; Byrne, C.D. Differential priming of RNA templates during cDNA synthesis markedly affects both accuracy and reproducibility of quantitative competitive reverse-transcriptase PCR. Biochem. J. 1999, 337, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Fellows, I. Deducer: A data analysis GUI for R. J. Stat. Softw. 2012, 49, 1–15. [Google Scholar] [CrossRef]


| Gene Symbol | HGNC 1 Approved Name | HGNC ID 2 | Location | Accession Number |
|---|---|---|---|---|
| ACTB | Actin β | 132 | 7p22.1 | NM_001101 |
| DDX17 | DEAD-box helicase 17 | 2740 | 22q13.1 | NM_001098504 |
| EXOC8 | Exocyst complex component 8 | 24,659 | 1q42.2 | NM_175876 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 4141 | 12p13.31 | NM_002046, NM_001289745, NM_001289746 |
| PPP1R15B | Protein phosphatase 1 regulatory subunit 15B | 14,951 | 1q32.1 | NM_032833 |
| RPL37A | Ribosomal protein L37a | 10,348 | 2q35 | NM_000998 |
| TPT1 | Tumor protein, translationally-controlled 1 | 12,022 | 13q14.13 | NM_001286272, NM_003295, NM_001286273 |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase | 30,892 | Xp11.22 | NM_031407 |
| Gene Exon | Exon ID (Ensembl, GRCh38.p10) | Mean RNA Levels (Counts) | SD (Counts) | CV |
|---|---|---|---|---|
| DDX17e1 | ENSE00001855680 | 1.3 | 0.52 | 40% |
| DDX17e2 | ENSE00001863979 | 1.6 | 0.55 | 34% |
| DDX17e3 | ENSE00001924900 | 66 | 9.4 | 14% |
| DDX17e4 | ENSE00001942031 | 85 | 6.6 | 8% |
| PPP1R15Be1 | ENSE00001443770 | 27 | 2.6 | 9% |
| PPP1R15Be2 | ENSE00001443771 | 43 | 4.2 | 10% |
| TPT1e1 | ENSE00001479630 | 13 | 3.6 | 28% |
| TPT1e2 | ENSE00001618156 | 63 | 13 | 21% |
| TPT1e3 | ENSE00001820001 | 272 | 74 | 27% |
| TPT1e4 | ENSE00001824735 | 0.45 | 0.69 | 153% |
| Patients Ending in sPTB 1 (n) | Patients Ending in TB 2 (n) | Total (n) | |
|---|---|---|---|
| Subset #1 | 9 | 8 | 17 |
| Subset #2 | 8 | 7 | 15 |
| Entire set | 17 | 15 | 32 |
| Gene | geNorm Stability Value | NormFinder Stability Value | ||||
|---|---|---|---|---|---|---|
| Rank in Set #1 | Rank in Set #2 | Absolute Rank Difference | Rank in Set #1 | Rank in Set #2 | Absolute Rank Difference | |
| ACTB | 2 | 1 | 1 | 3 | 2 | 1 |
| DDX17 | 5 | 5 | 0 | 6 | 5 | 1 |
| EXOC8 | 3 | 3 | 0 | 1 | 1 | 0 |
| GAPDH | 6 | 6 | 0 | 5 | 6 | 1 |
| PPP1R15B | 1 | 2 | 1 | 2 | 3 | 1 |
| TPT1 | 4 | 4 | 0 | 4 | 4 | 0 |
| Total | - | - | 2 | - | - | 4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chim, S.S.C.; Wong, K.K.W.; Chung, C.Y.L.; Lam, S.K.W.; Kwok, J.S.L.; Lai, C.-Y.; Cheng, Y.K.Y.; Hui, A.S.Y.; Meng, M.; Chan, O.-K.; et al. Systematic Selection of Reference Genes for the Normalization of Circulating RNA Transcripts in Pregnant Women Based on RNA-Seq Data. Int. J. Mol. Sci. 2017, 18, 1709. https://doi.org/10.3390/ijms18081709
Chim SSC, Wong KKW, Chung CYL, Lam SKW, Kwok JSL, Lai C-Y, Cheng YKY, Hui ASY, Meng M, Chan O-K, et al. Systematic Selection of Reference Genes for the Normalization of Circulating RNA Transcripts in Pregnant Women Based on RNA-Seq Data. International Journal of Molecular Sciences. 2017; 18(8):1709. https://doi.org/10.3390/ijms18081709
Chicago/Turabian StyleChim, Stephen S. C., Karen K. W. Wong, Claire Y. L. Chung, Stephanie K. W. Lam, Jamie S. L. Kwok, Chit-Ying Lai, Yvonne K. Y. Cheng, Annie S. Y. Hui, Meng Meng, Oi-Ka Chan, and et al. 2017. "Systematic Selection of Reference Genes for the Normalization of Circulating RNA Transcripts in Pregnant Women Based on RNA-Seq Data" International Journal of Molecular Sciences 18, no. 8: 1709. https://doi.org/10.3390/ijms18081709