Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes
Abstract
:1. Introduction
2. Photoreceptors as Molecular Templates for Engineering of Fluorescent Proteins
2.1. Structure and Photoconversion of Bacterial Phytochromes
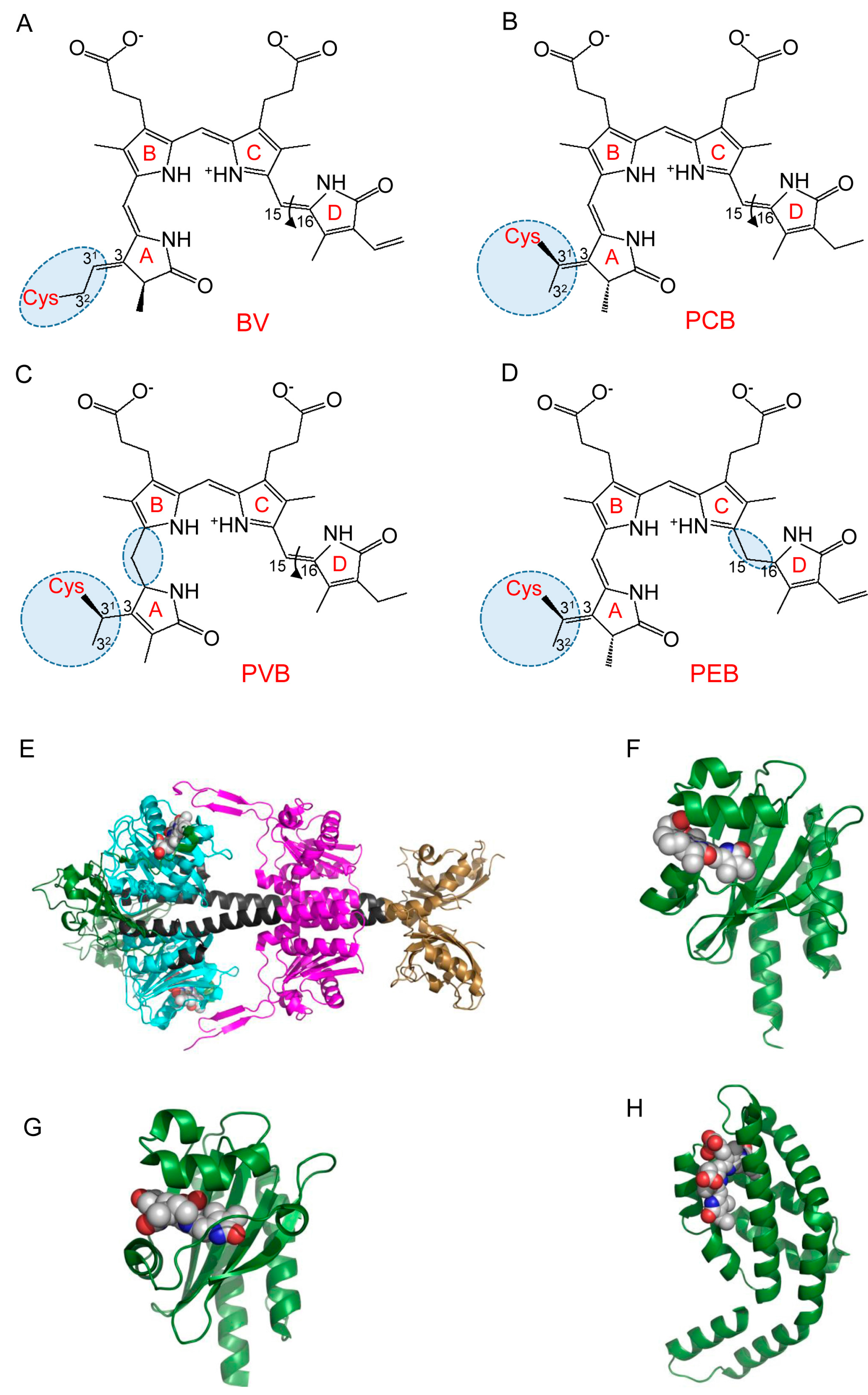
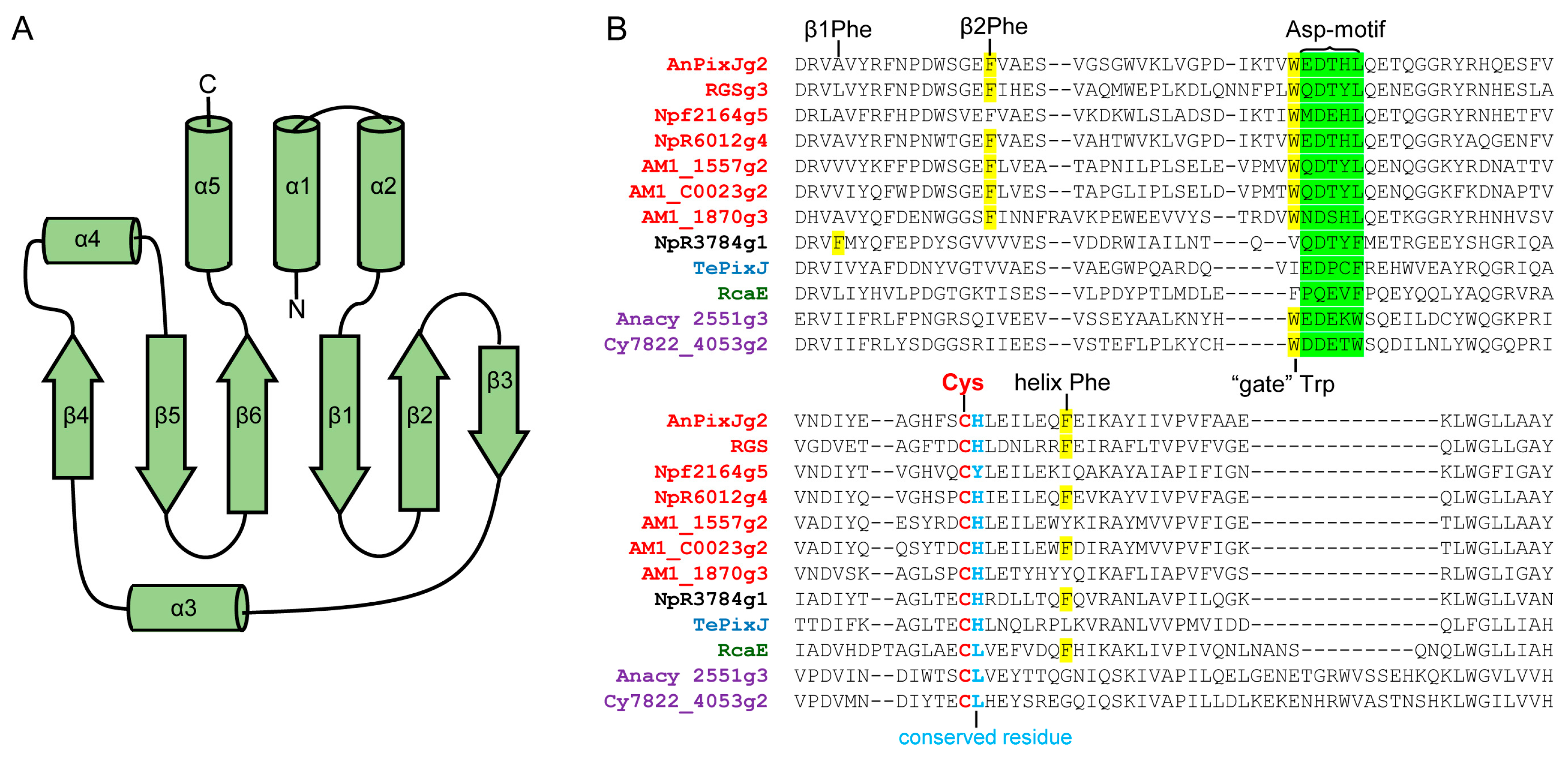
2.2. Structure and Properties of Cyanobacteriochromes
2.3. Allophycocyanins as Source of FPs
3. Engineering Approaches to Develop Near-Infrared Fluorescent Proteins
3.1. Examples of Rational Design of Near-Infrared Fluorescent Proteins
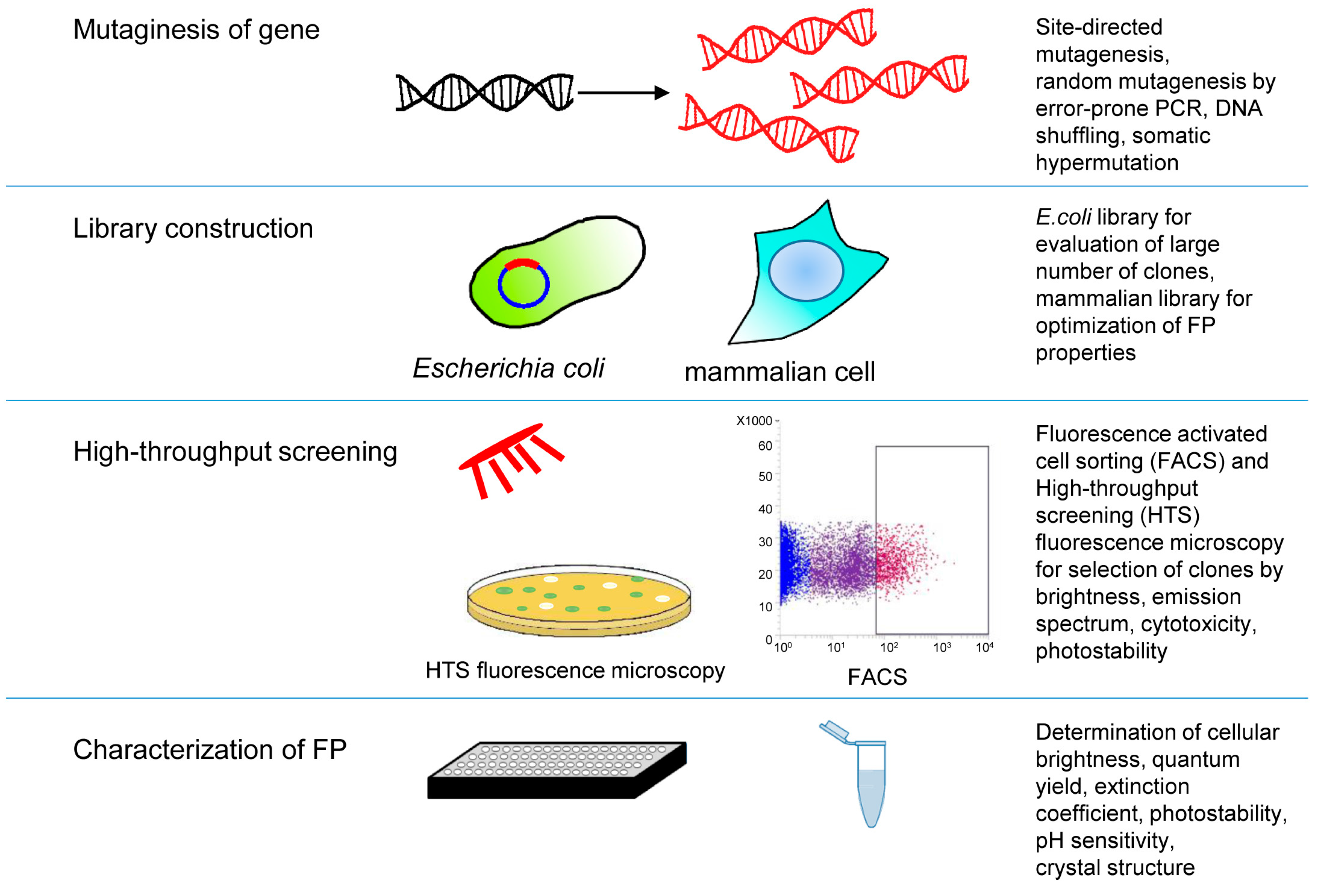
3.2. High-Throughput Screening Approaches of Near-Infrared Fluorescent Proteins
4. Biological Applications of Near-Infrared Fluorescent Proteins
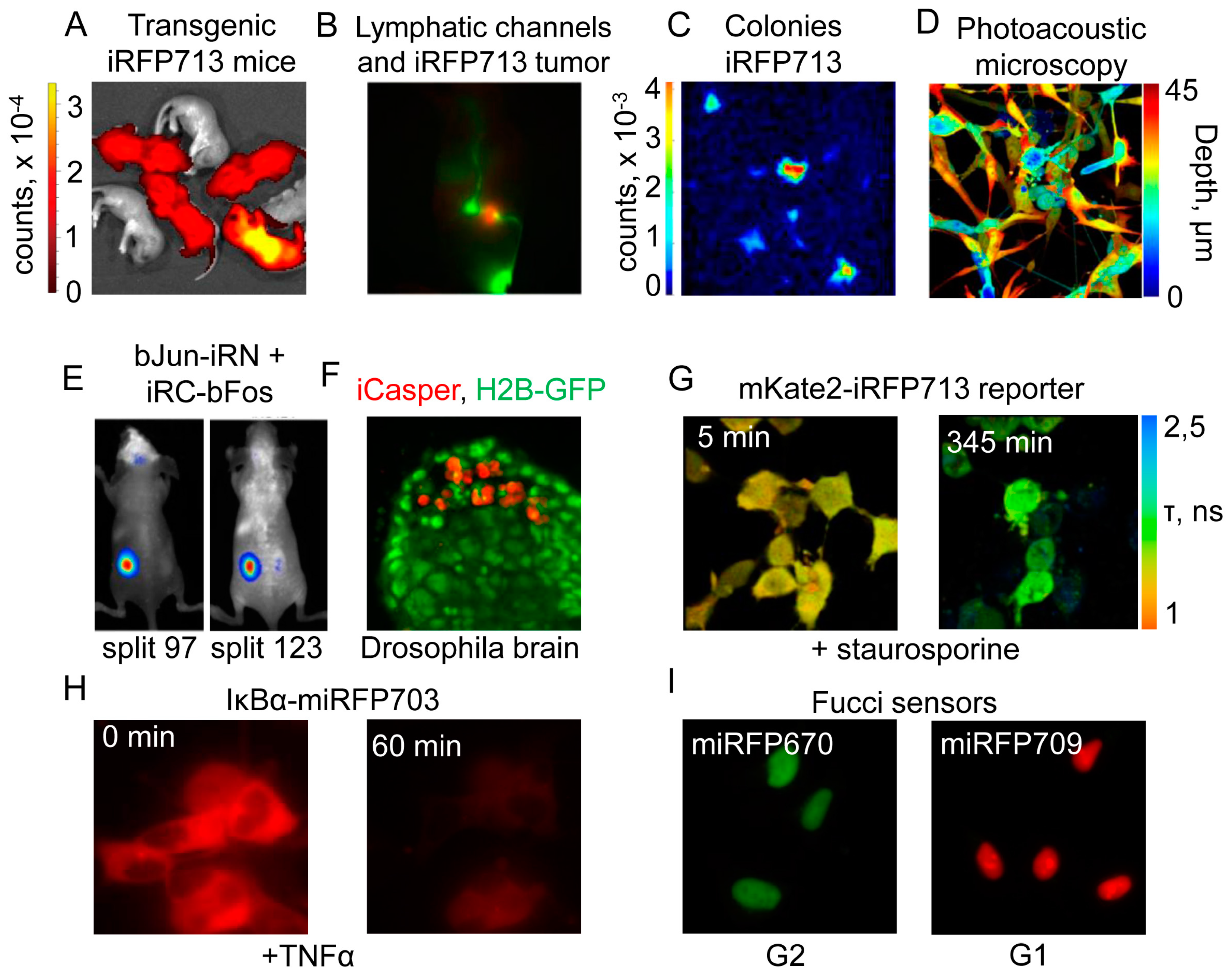
4.1. Application of Near-Infrared Fluorescent Proteins to In Vivo Imaging
4.2. Near-Infrared Fluorescent Proteins in Cancer Research
4.3. Near-Infrared Imaging in High-Throughput Applications
4.4. Near-Infrared Fluorescent Proteins in Microbiology and Parasitology
4.5. Near-Infrared Fluorescent Proteins in Advanced Imaging Techniques
4.6. Biosensors and Reporters Designed from Near-Infrared Fluorescent Proteins
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| APC | Allophycocyanin |
| BphP | Bacterial phytochrome photoreceptor |
| BV | Biliverdin |
| CBCR | Cyanobacteriochrome |
| FP | Fluorescent protein |
| HO | Heme oxygenase |
| iRFP | Near-infraRed Fluorescent Protein |
| miRFP | Monomeric near-infraRed Fluorescent Protein |
| NIR | Near-infrared |
| PCB | Phycocyanobilin |
| PCM | Photosensory core module |
| PEB | Phycoerythrobilin |
| PPIX | Protoporphyrin IX |
| PΦB | Phytochromobilin |
| PVB | Phycoviolobilin |
References
- Shcherbakova, D.M.; Subach, O.M.; Verkhusha, V.V. Red fluorescent proteins: Advanced imaging applications and future design. Angew. Chem. 2012, 51, 10724–10738. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Shemetov, A.A.; Kaberniuk, A.A.; Verkhusha, V.V. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu. Rev. Biochem. 2015, 84, 519–550. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Subach, F.V.; Verkhusha, V.V. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 2013, 42, 3441–3452. [Google Scholar] [CrossRef] [PubMed]
- Chernov, K.G.; Redchuk, T.A.; Omelina, E.S.; Verkhusha, V.V. Near-infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 2017, 117, 6423–6446. [Google Scholar] [CrossRef] [PubMed]
- Drobizhev, M.; Makarov, N.S.; Tillo, S.E.; Hughes, T.E.; Rebane, A. Two-photon absorption properties of fluorescent proteins. Nat. Methods 2011, 8, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Verkhusha, V.V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Shemetov, A.A.; Oliinyk, O.S.; Verkhusha, V.V. How to increase brightness of near-infrared fluorescent proteins in mammalian cells. Cell Chem. Biol. 2017, 24, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Redchuk, T.A.; Omelina, E.S.; Chernov, K.G.; Verkhusha, V.V. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat. Chem. Biol. 2017, 13, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Telford, W.G.; Shcherbakova, D.M.; Buschke, D.; Hawley, T.S.; Verkhusha, V.V. Multiparametric flow cytometry using near-infrared fluorescent proteins engineered from bacterial phytochromes. PLoS ONE 2015, 10, e0122342. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.W.; Chen, H.L.; Yen, C.C.; Wang, J.L.; Yang, S.H.; Chen, C.M. Using dual fluorescence reporting genes to establish an in vivo imaging model of orthotopic lung adenocarcinoma in mice. Mol. Imaging Biol. 2016, 18, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Rumyantsev, K.A.; Shcherbakova, D.M.; Zakharova, N.I.; Emelyanov, A.V.; Turoverov, K.K.; Verkhusha, V.V. Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence. Sci. Rep. 2015, 5, 18348. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Smith, H. Phytochromes and light signal perception by plants—An emerging synthesis. Nature 2000, 407, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011, 21, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Verméglio, A. Bacteriophytochromes in anoxygenic photosynthetic bacteria. Photosynth. Res. 2008, 97, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Anders, K.; Essen, L.O. The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 2015, 35, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Vierstra, R.D. Phytochromes: An atomic perspective on photoactivation and signaling. Plant Cell 2014, 26, 4568–4583. [Google Scholar] [CrossRef] [PubMed]
- Bellini, D.; Papiz, M.Z. Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 2012, 20, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Bjorling, A.; Linna, M.; Westenhoff, S.; Ihalainen, J.A. Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem. 2015, 290, 16383–16392. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Björling, A.; Berntsson, O.; Lehtivuori, H.; Niebling, S.; Hoernke, M.; Kosheleva, I.; Henning, R.; Menzel, A.; Ihalainen, J.A.; et al. Signal amplification and transduction in phytochrome photosensors. Nature 2014, 509, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.L.; Zhulin, I.B. Pas domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [PubMed]
- Möglich, A.; Ayers, R.A.; Moffat, K. Structure and signaling mechanism of per-arnt-sim domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef] [PubMed]
- Moglich, A.; Yang, X.; Ayers, R.A.; Moffat, K. Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 2010, 61, 21–47. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Zhang, J.; Vierstra, R.D. Crystal structure of deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 2016, 24, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.T.; Tanaka, J.; Hamada, M.; Sugiyama, Y.; Sakaguchi, S.; Nakamura, M.; Takahashi, S.; Miwa, Y. In vivo image analysis using irfp transgenic mice. Exp. Anim. Jpn. Assoc. Lab. Anim. Sci. 2014, 63, 311–319. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Pletnev, S.; Malashkevich, V.N.; Xiao, H.; Dauter, Z.; Verkhusha, V.V. Molecular basis of spectral diversity in near-infrared phytochrome-based fluorescent proteins. Chem. Biol. 2015, 22, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Karniol, B.; Vierstra, R.D. The pair of bacteriophytochromes from agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 2003, 100, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.J.; Lagarias, J.C. Harnessing phytochrome’s glowing potential. Proc. Natl. Acad. Sci. USA 2004, 101, 17334–17339. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Zhang, J.; von Stetten, D.; Gunther, M.; Murgida, D.H.; Mroginski, M.A.; Walker, J.M.; Forest, K.T.; Hildebrandt, P.; Vierstra, R.D. Mutational analysis of deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome. Proc. Natl. Acad. Sci. USA 2010, 107, 9170–9175. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Royant, A.; Lin, M.Z.; Aguilera, T.A.; Lev-Ram, V.; Steinbach, P.A.; Tsien, R.Y. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 2009, 324, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Maines, M.D. The heme oxygenase system: A regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 517–554. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011, 29, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Baird, M.A.; Allen, J.R.; Howe, E.S.; Klassen, M.P.; Reade, A.; Makhijani, K.; Song, Y.; Liu, S.; Murthy, Z.; et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 2015, 12, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.Y.; Soulier, N.T.; Canniffe, D.P.; Shen, G.; Bryant, D.A. Light regulation of pigment and photosystem biosynthesis in cyanobacteria. Curr. Opin. Plant Biol. 2017, 37, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Lagarias, J.C. A brief history of phytochromes. ChemPhysChem 2010, 11, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Ishizuka, T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008, 7, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Feoktistova, K.; Lagarias, J.C. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. USA 2011, 108, 11854–11859. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Mechanistic insight into the photosensory versatility of dxcf cyanobacteriochromes. Biochemistry 2012, 51, 3576–3585. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Rockwell, N.C.; Enomoto, G.; Win, N.-N.; Martin, S.S.; Gan, F.; Bryant, D.A.; Ikeuchi, M.; Lagarias, J.C.; Narikawa, R. Cyanobacteriochrome photoreceptors lacking the canonical cys residue. Biochemistry 2016, 55, 6981–6995. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Xu, J.G.; Tang, K.; Miao, D.; Gartner, W.; Scheer, H.; Zhao, K.H.; Zhou, M. Orange fluorescent proteins constructed from cyanobacteriochromes chromophorylated with phycoerythrobilin. Photochem. Photobiol. Sci. 2014, 13, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Nakajima, T.; Aono, Y.; Fushimi, K.; Enomoto, G.; Win, N.-N.; Itoh, S.; Sato, M.; Ikeuchi, M. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium acaryochloris marina. Sci. Rep. 2015, 5, 7950. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Fushimi, K.; Win, N.-N.; Ikeuchi, M. Red-shifted red/green-type cyanobacteriochrome am1_1870g3 from the chlorophyll d-bearing cyanobacterium acaryochloris marina. Biochem. Biophys. Res. Commun. 2015, 461, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Nakajima, T.; Aono, Y.; Yamamoto, T.; Win, N.-N.; Ikeuchi, M.; Sato, M.; Narikawa, R. Photoconversion and fluorescence properties of a red/green-type cyanobacteriochrome am1_c0023g2 that binds not only phycocyanobilin but also biliverdin. Front. Microbiol. 2016, 7, 588. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Identification of dxcf cyanobacteriochrome lineages with predictable photocycles. Photochem. Photobiol. Sci. 2015, 14, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Ishizuka, T.; Muraki, N.; Shiba, T.; Kurisu, G.; Ikeuchi, M. Structures of cyanobacteriochromes from phototaxis regulators anpixj and tepixj reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Walker, J.M.; Phillips, G.N., Jr.; Vierstra, R.D. A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in dxcf-cyanobacteriochromes. Structure 2013, 21, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Cornilescu, C.C.; Cornilescu, G.; Burgie, E.S.; Markley, J.L.; Ulijasz, A.T.; Vierstra, R.D. Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 2014, 289, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Zhang, J.; Brunzelle, J.S.; Vierstra, R.D.; Forest, K.T. High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 2007, 282, 12298–12309. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Suzuki, F.; Yoshihara, S.; Higashi, S.; Watanabe, M.; Ikeuchi, M. Novel photosensory two-component system (pixa-nixb-nixc) involved in the regulation of positive and negative phototaxis of cyanobacterium synechocystis sp. Pcc 6803. Plant Cell Physiol. 2011, 52, 2214–2224. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Kohchi, T.; Ikeuchi, M. Characterization of the photoactive gaf domain of the cika homolog (sycika, slr1969) of the cyanobacterium Synechocystis sp. Pcc 6803. Photochem. Photobiol. Sci. 2008, 7, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Phycoviolobilin formation and spectral tuning in the dxcf cyanobacteriochrome subfamily. Biochemistry 2012, 51, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Wiltbank, L.B.; Kehoe, D.M. Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow, and red light. MBio 2016, 7, e02130-15. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Enomoto, G.; Win, N.-N.; Fushimi, K.; Ikeuchi, M. A new type of dual-cys cyanobacteriochrome gaf domain found in cyanobacterium acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 2014, 53, 5051–5059. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Rockwell, N.C.; Nishiyama, K.; Narikawa, R.; Ukaji, Y.; Inomata, K.; Lagarias, J.C.; Ikeuchi, M. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. USA 2013, 110, 4974–4979. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Identification of cyanobacteriochromes detecting far-red light. Biochemistry 2016, 55, 3907–3919. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Kamiya, A.; Suzuki, H.; Narikawa, R.; Noguchi, T.; Kohchi, T.; Inomata, K.; Ikeuchi, M. The cyanobacteriochrome, tepixj, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 2011, 50, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Narikawa, R.; Kohchi, T.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome tepixj of thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007, 48, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 2014, 53, 3118–3130. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, D.M.; Gutu, A. Responding to color: The regulation of complementary chromatic adaptation. Annu. Rev. Plant Biol. 2006, 57, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Gutu, A.; Kehoe, D.M. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 2012, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, K.; Montgomery, B.L.; Grossman, A.R.; Lagarias, J.C.; Kehoe, D.M. Rcae is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol. Microbiol. 2004, 51, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome ccas regulates phycoerythrin accumulation in nostoc punctiforme, a group ii chromatic adapter. Proc. Natl. Acad. Sci. USA 2010, 107, 8854–8859. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Shimada, T.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome ccas is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. USA 2008, 105, 9528–9533. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Abe, K.; Ferri, S.; Sode, K. Development of a light-regulated cell-recovery system for non-photosynthetic bacteria. Microb. Cell Fact. 2016, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Tabor, J.J.; Levskaya, A.; Voigt, C.A. Multichromatic control of gene expression in escherichia coli. J. Mol. Biol. 2011, 405, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Miyake, K.; Nakamura, M.; Kojima, K.; Ferri, S.; Ikebukuro, K.; Sode, K. Engineering of a green-light inducible gene expression system in Synechocystis sp. Pcc6803. Microb. Biotechnol. 2014, 7, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Rockwell, N.C.; Martin, S.S.; Dallas, J.L.; Lagarias, J.C.; Ames, J.B. Photoconversion changes bilin chromophore conjugation and protein secondary structure in the violet/orange cyanobacteriochrome npf2164g3′ [corrected]. Photochem. Photobiol. Sci. 2014, 13, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Njuguna, S.L.; Roberts, L.; Castillo, E.; Parson, V.L.; Dwojak, S.; Lagarias, J.C.; Spiller, S.C. A second conserved gaf domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome tlr0924 from thermosynechococcus elongatus. Biochemistry 2008, 47, 7304–7316. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; Jeoung, S.C.; Song, J.Y.; Song, J.J.; Park, Y.I. Hydrophobic residues near the bilin chromophore-binding pocket modulate spectral tuning of insert-cys subfamily cyanobacteriochromes. Sci. Rep. 2017, 7, 40576. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; Jeoung, S.C.; Song, J.Y.; Kupriyanova, E.V.; Pronina, N.A.; Lee, B.W.; Jo, S.W.; Park, B.S.; Choi, S.B.; Song, J.J.; et al. Genomic survey and biochemical analysis of recombinant candidate cyanobacteriochromes reveals enrichment for near uv/violet sensors in the halotolerant and alkaliphilic cyanobacterium microcoleus ippas b353. J. Biol. Chem. 2015, 290, 28502–28514. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Tabor, J.J. Repurposing Synechocystis pcc6803 uirs-uirr as a uv-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth. Biol. 2016, 5, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry 2012, 51, 9667–9677. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Fukushima, Y.; Ishizuka, T.; Itoh, S.; Ikeuchi, M. A novel photoactive gaf domain of cyanobacteriochrome anpixj that shows reversible green/red photoconversion. J. Mol. Biol. 2008, 380, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Velazquez Escobar, F.; Utesch, T.; Narikawa, R.; Ikeuchi, M.; Mroginski, M.A.; Gartner, W.; Hildebrandt, P. Photoconversion mechanism of the second gaf domain of cyanobacteriochrome anpixj and the cofactor structure of its green-absorbing state. Biochemistry 2013, 52, 4871–4880. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Velazquez Escobar, F.; Xu, X.L.; Narikawa, R.; Ikeuchi, M.; Siebert, F.; Gartner, W.; Matysik, J.; Hildebrandt, P. A red/green cyanobacteriochrome sustains its color despite a change in the bilin chromophore’s protonation state. Biochemistry 2015, 54, 5839–5848. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Narikawa, R.; Ikeuchi, M.; Gartner, W.; Matysik, J. Color tuning in red/green cyanobacteriochrome anpixj: Photoisomerization at c15 causes an excited-state destabilization. J. Phys. Chem. B 2015, 119, 9688–9695. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, S.M.; Kim, P.W.; Chang, C.W.; Hanke, S.J.; Hayer, R.J.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Larsen, D.S. Conservation and diversity in the primary forward photodynamics of red/green cyanobacteriochromes. Biochemistry 2015, 54, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Gan, F.; Bryant, D.A.; Lagarias, J.C. Npr3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 2015, 14, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Slavov, C.; Xu, X.; Zhao, K.H.; Gartner, W.; Wachtveitl, J. Detailed insight into the ultrafast photoconversion of the cyanobacteriochrome slr1393 from Synechocystis sp. Biochim. Biophys. Acta 2015, 1847, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, X.J.; Wang, Z.B.; Chen, Y.; Wang, X.; Zhou, M.; Scheer, H.; Zhao, K.H. Fused-gene approach to photoswitchable and fluorescent biliproteins. Angew. Chem. 2010, 49, 5456–5458. [Google Scholar] [CrossRef] [PubMed]
- Pennacchietti, F.; Losi, A.; Xu, X.L.; Zhao, K.H.; Gartner, W.; Viappiani, C.; Cella, F.; Diaspro, A.; Abbruzzetti, S. Photochromic conversion in a red/green cyanobacteriochrome from Synechocystis pcc6803: Quantum yields in solution and photoswitching dynamics in living e. Coli cells. Photochem. Photobiol. Sci. 2015, 14, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Gustafson, W.C.; Han, C.; Lafaye, C.; Noirclerc-Savoye, M.; Ge, W.P.; Thayer, D.A.; Huang, H.; Kornberg, T.B.; Royant, A.; et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014, 5, 3626. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Subach, F.V.; Verkhusha, V.V. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 2013, 4, 2153. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lim, S.; Lagarias, J.C.; Ames, J.B. Characterization of red/green cyanobacteriochrome npr6012g4 by solution nuclear magnetic resonance spectroscopy: A hydrophobic pocket for the c15-e,anti chromophore in the photoproduct. Biochemistry 2015, 54, 3772–3783. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Ding, W.L.; Hoppner, A.; Zhao, C.; Zhang, L.; Hontani, Y.; Kennis, J.T.; Gartner, W.; Scheer, H.; Zhou, M.; et al. The terminal phycobilisome emitter, lcm: A light-harvesting pigment with a phytochrome chromophore. Proc. Natl. Acad. Sci. USA 2015, 112, 15880–15885. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.P.; Dong, L.L.; Sun, Y.F.; Zeng, X.L.; Ding, W.L.; Scheer, H.; Yang, X.; Zhao, K.H. The structure of allophycocyanin b from Synechocystis pcc 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.R.; Schaefer, M.R.; Chiang, G.G.; Collier, J.L. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 1993, 57, 725–749. [Google Scholar] [PubMed]
- Trinquet, E.; Maurin, F.; Preaudat, M.; Mathis, G. Allophycocyanin 1 as a near-infrared fluorescent tracer: Isolation, characterization, chemical modification, and use in a homogeneous fluorescence resonance energy transfer system. Anal. Biochem. 2001, 296, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Loos, D.; Cotlet, M.; De Schryver, F.; Habuchi, S.; Hofkens, J. Single-molecule spectroscopy selectively probes donor and acceptor chromophores in the phycobiliprotein allophycocyanin. Biophys. J. 2004, 87, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Batard, P.; Szollosi, J.; Luescher, I.; Cerottini, J.-C.; MacDonald, R.; Romero, P. Use of phycoerythrin and allophycocyanin for fluorescence resonance energy transfer analyzed by flow cytometry: Advantages and limitations. Cytometry 2002, 48, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Vasquez, Y.M.; Dragomani, T.M.; Kronfel, M.L.; Williams, S.R.; Alvey, R.M.; Bryant, D.A.; Schluchter, W.M. Biosynthesis of cyanobacterial phycobiliproteins in escherichia coli: Chromophorylation efficiency and specificity of all bilin lyases from synechococcus sp. Strain pcc 7002. Appl. Environ. Microbiol. 2010, 76, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Hu, I.C.; Lee, T.R.; Lin, H.F.; Chiueh, C.C.; Lyu, P.C. Biosynthesis of fluorescent allophycocyanin alpha-subunits by autocatalytic bilin attachment. Biochemistry 2006, 45, 7092–7099. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.Z.; Han, J.X.; Tang, Q.Y.; Ding, W.L.; Miao, D.; Zhou, M.; Scheer, H.; Zhao, K.H. Far-red light photoacclimation: Chromophorylation of fr induced alpha- and beta-subunits of allophycocyanin from chroococcidiopsis thermalis sp. Pcc7203. Biochim. Biophys. Acta 2016, 1857, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Sonani, R.R.; Gupta, G.D.; Madamwar, D.; Kumar, V. Crystal structure of allophycocyanin from marine cyanobacterium phormidium sp. A09dm. PLoS ONE 2015, 10, e0124580. [Google Scholar] [CrossRef] [PubMed]
- Brejc, K.; Ficner, R.; Huber, R.; Steinbacher, S. Isolation, crystallization, crystal structure analysis and refinement of allophycocyanin from the cyanobacterium spirulina platensis at 2.3 a resolution. J. Mol. Biol. 1995, 249, 424–440. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Jiang, T.; Zhang, J.P.; Liang, D.C. Crystal structure of allophycocyanin from red algae porphyra yezoensis at 2.2-a resolution. J. Biol. Chem. 1999, 274, 16945–16952. [Google Scholar] [CrossRef] [PubMed]
- Scheer, H.; Zhao, K.H. Biliprotein maturation: The chromophore attachment. Mol. Microbiol. 2008, 68, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.A.; Tran, G.N.; Gross, L.A.; Crisp, J.L.; Shu, X.; Lin, J.Y.; Tsien, R.Y. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 2016, 13, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Miao, D.; Ding, W.L.; Zhao, B.Q.; Lu, L.; Xu, Q.Z.; Scheer, H.; Zhao, K.H. Adapting photosynthesis to the near-infrared: Non-covalent binding of phycocyanobilin provides an extreme spectral red-shift to phycobilisome core-membrane linker from synechococcus sp. Pcc7335. Biochim. Biophys. Acta 2016, 1857, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, T.; Moglich, A. Photoreceptor engineering. Front. Mol. Biosci. 2015, 2, 30. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Ponka, P. Cell biology of heme. Am. J. Med. Sci. 1999, 318, 241–256. [Google Scholar] [CrossRef]
- Lehtivuori, H.; Bhattacharya, S.; Angenent-Mari, N.M.; Satyshur, K.A.; Forest, K.T. Removal of chromophore-proximal polar atoms decreases water content and increases fluorescence in a near infrared phytofluor. Front. Mol. Biosci. 2015, 2, 65. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs. Phys. Chem. Chem. Phys. 2011, 13, 11985–11997. [Google Scholar] [CrossRef] [PubMed]
- Zienicke, B.; Chen, L.Y.; Khawn, H.; Hammam, M.A.; Kinoshita, H.; Reichert, J.; Ulrich, A.S.; Inomata, K.; Lamparter, T. Fluorescence of phytochrome adducts with synthetic locked chromophores. J. Biol. Chem. 2011, 286, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Hontani, Y.; Shcherbakova, D.M.; Baloban, M.; Zhu, J.; Verkhusha, V.V.; Kennis, J.T. Bright blue-shifted fluorescent proteins with cys in the gaf domain engineered from bacterial phytochromes: Fluorescence mechanisms and excited-state dynamics. Sci. Rep. 2016, 6, 37362. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Baloban, M.; Bublikov, G.S.; Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Kuznetsova, I.M.; Verkhusha, V.V. Allosteric effects of chromophore interaction with dimeric near-infrared fluorescent proteins engineered from bacterial phytochromes. Sci. Rep. 2016, 6, 18750. [Google Scholar] [CrossRef] [PubMed]
- Rumyantsev, K.A.; Shcherbakova, D.M.; Zakharova, N.I.; Verkhusha, V.V.; Turoverov, K.K. Design of near-infrared single-domain fluorescent protein gaf-fp based on bacterial phytochrome. Cell Tissue Biol. 2017, 11, 16–26. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Satyshur, K.A.; Anstrom, D.M.; Forest, K.T. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 2012, 287, 7000–7009. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K. Interaction of biliverdin chromophore with near-infrared fluorescent protein bphp1-fp engineered from bacterial phytochrome. Int. J. Mol. Sci. 2017, 18, 1009. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shcherbakova, D.M.; Hontani, Y.; Verkhusha, V.V.; Kennis, J.T. Ultrafast excited-state dynamics and fluorescence deactivation of near-infrared fluorescent proteins engineered from bacteriophytochromes. Sci. Rep. 2015, 5, 12840. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Auldridge, M.E.; Lehtivuori, H.; Ihalainen, J.A.; Forest, K.T. Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 2014, 289, 32144–32152. [Google Scholar] [CrossRef] [PubMed]
- Buhrke, D.; Velazquez Escobar, F.; Sauthof, L.; Wilkening, S.; Herder, N.; Tavraz, N.N.; Willoweit, M.; Keidel, A.; Utesch, T.; Mroginski, M.A.; et al. The role of local and remote amino acid substitutions for optimizing fluorescence in bacteriophytochromes: A case study on irfp. Sci. Rep. 2016, 6, 28444. [Google Scholar] [CrossRef] [PubMed]
- Miralem, T.; Lerner-Marmarosh, N.; Gibbs, P.E.; Tudor, C.; Hagen, F.K.; Maines, M.D. The human biliverdin reductase-based peptide fragments and biliverdin regulate protein kinase cdelta activity: The peptides are inhibitors or substrate for the protein kinase c. J. Biol. Chem. 2012, 287, 24698–24712. [Google Scholar] [CrossRef] [PubMed]
- Wegiel, B.; Baty, C.J.; Gallo, D.; Csizmadia, E.; Scott, J.R.; Akhavan, A.; Chin, B.Y.; Kaczmarek, E.; Alam, J.; Bach, F.H.; et al. Cell surface biliverdin reductase mediates biliverdin-induced anti-inflammatory effects via phosphatidylinositol 3-kinase and akt. J. Biol. Chem. 2009, 284, 21369–21378. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.; Tampe, J.; Shing, C.; Bakrania, B.; Winearls, J.; Fraser, J.; Wagner, K.H.; Bulmer, A.C. Endogenous tetrapyrroles influence leukocyte responses to lipopolysaccharide in human blood: Pre-clinical evidence demonstrating the anti-inflammatory potential of biliverdin. J. Clin. Cell Immunol. 2014, 5, 1000218. [Google Scholar] [PubMed]
- Gibbs, P.E.; Maines, M.D. Biliverdin inhibits activation of nf-kappab: Reversal of inhibition by human biliverdin reductase. Int. J. Cancer 2007, 121, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Phelan, D.; Winter, G.M.; Rogers, W.J.; Lam, J.C.; Denison, M.S. Activation of the ah receptor signal transduction pathway by bilirubin and biliverdin. Arch. Biochem. Biophys. 1998, 357, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Cirino, P.C.; Mayer, K.M.; Umeno, D. Generating mutant libraries using error-prone pcr. Methods Mol. Biol. 2003, 231, 3–9. [Google Scholar] [PubMed]
- Wilson, D.S.; Keefe, A.D. Random mutagenesis by pcr. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Shafikhani, S.; Siegel, R.A.; Ferrari, E.; Schellenberger, V. Generation of large libraries of random mutants in bacillus subtilis by pcr-based plasmid multimerization. Biotechniques 1997, 23, 304–310. [Google Scholar] [PubMed]
- Martinez, R.; Schwaneberg, U. A roadmap to directed enzyme evolution and screening systems for biotechnological applications. Biol. Res. 2013, 46, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Hanson-Manful, P.; Patrick, W.M. Construction and analysis of randomized protein-encoding libraries using error-prone pcr. Methods Mol. Biol. 2013, 996, 251–267. [Google Scholar] [PubMed]
- Wong, T.S.; Tee, K.L.; Hauer, B.; Schwaneberg, U. Sequence saturation mutagenesis (sesam): A novel method for directed evolution. Nucleic Acids Res. 2004, 32, e26. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Esteban, O.; Woodyer, R.D.; Zhao, H. In vitro DNA recombination by random priming. Methods Mol. Biol. 2003, 231, 99–104. [Google Scholar] [PubMed]
- Drummond, D.A.; Iverson, B.L.; Georgiou, G.; Arnold, F.H. Why high-error-rate random mutagenesis libraries are enriched in functional and improved proteins. J. Mol. Biol. 2005, 350, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Dean, K.M.; Lubbeck, J.L.; Davis, L.M.; Regmi, C.K.; Chapagain, P.P.; Gerstman, B.S.; Jimenez, R.; Palmer, A.E. Microfluidics-based selection of red-fluorescent proteins with decreased rates of photobleaching. Integr. Biol. 2015, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, J.; Schnitzer, M.J. An infrared fluorescent protein for deeper imaging. Nat. Biotechnol. 2011, 29, 715–716. [Google Scholar] [CrossRef] [PubMed]
- Rumyantsev, K.A.; Turoverov, K.K.; Verkhusha, V.V. Near-infrared bioluminescent proteins for two-color multimodal imaging. Sci. Rep. 2016, 6, 36588. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Krumholz, A.; Xia, J.; Yao, J.; Wang, L.V.; Verkhusha, V.V. Deep-tissue photoacoustic tomography of a genetically encoded near-infrared fluorescent probe. Angew. Chem. 2012, 51, 1448–1451. [Google Scholar] [CrossRef] [PubMed]
- Fyk-Kolodziej, B.; Hellmer, C.B.; Ichinose, T. Marking cells with infrared fluorescent proteins to preserve photoresponsiveness in the retina. Biotechniques 2014, 57, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, M.; Wang, X.; Qin, G.; Weintraub, N.L.; Tang, Y. Assessing in vitro stem-cell function and tracking engraftment of stem cells in ischaemic hearts by using novel irfp gene labelling. J. Cell Mol. Med. 2014, 18, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Hock, A.K.; Cheung, E.C.; Humpton, T.J.; Monteverde, T.; Paulus-Hock, V.; Lee, P.; McGhee, E.; Scopelliti, A.; Murphy, D.J.; Strathdee, D.; et al. Development of an inducible mouse model of irfp713 to track recombinase activity and tumour development in vivo. Sci. Rep. 2017, 7, 1837. [Google Scholar] [CrossRef] [PubMed]
- Agollah, G.D.; Wu, G.; Sevick-Muraca, E.M.; Kwon, S. In vivo lymphatic imaging of a human inflammatory breast cancer model. J. Cancer 2014, 5, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Paulus-Hock, V.; Cheung, E.C.; Roxburgh, P.; Vousden, K.H.; Hock, A.K. iRFP is a real time marker for transformation based assays in high content screening. PLoS ONE 2014, 9, e98399. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Kaberniuk, A.A.; Li, L.; Shcherbakova, D.M.; Zhang, R.; Wang, L.; Li, G.; Verkhusha, V.V.; Wang, L.V. Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat. Methods 2016, 13, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, W.; Zhang, Z.; Liu, S.; Zhang, X.; Zhang, X.E.; Cui, Z. Novel near-infrared bifc systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions. Biomaterials 2015, 48, 97–107. [Google Scholar] [CrossRef] [PubMed]
- To, T.L.; Piggott, B.J.; Makhijani, K.; Yu, D.; Jan, Y.N.; Shu, X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. USA 2015, 112, 3338–3343. [Google Scholar] [CrossRef] [PubMed]
- Zlobovskaya, O.A.; Sergeeva, T.F.; Shirmanova, M.V.; Dudenkova, V.V.; Sharonov, G.V.; Zagaynova, E.V.; Lukyanov, K.A. Genetically encoded far-red fluorescent sensors for caspase-3 activity. Biotechniques 2016, 60, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Jiguet-Jiglaire, C.; Cayol, M.; Mathieu, S.; Jeanneau, C.; Bouvier-Labit, C.; Ouafik, L.; El-Battari, A. Noninvasive near-infrared fluorescent protein-based imaging of tumor progression and metastases in deep organs and intraosseous tissues. J. Biomed. Opt. 2014, 19, 16019. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Robinson, H.; Zhang, S.; Wu, G.; Sevick-Muraca, E.M. Longitudinal far red gene-reporter imaging of cancer metastasis in preclinical models: A tool for accelerating drug discovery. Biomed. Opt. Express 2015, 6, 3346–3351. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.H.; Wu, G.; Robinson, H.; Wilganowski, N.; Hall, M.A.; Ghosh, S.C.; Pinkston, K.L.; Azhdarinia, A.; Harvey, B.R.; Sevick-Muraca, E.M. Tumor margin detection using quantitative nirf molecular imaging targeting epcam validated by far red gene reporter irfp. Mol. Imaging Biol. 2013, 15, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Strukelj, B. A high-throughput biliverdin assay using infrared fluorescence. J. Vet. Diagn. Investig. 2014, 26, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Zhao, M.; Sheng, Y.; Bentolila, L.A.; Tang, Y. Detection of mercury ion by infrared fluorescent protein and its hydrogel-based paper assay. Anal. Chem. 2011, 83, 2324–2329. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Zavrsnik, J.; Butinar, M.; Turk, B.; Strukelj, B. In vivo imaging of lactococcus lactis, lactobacillus plantarum and escherichia coli expressing infrared fluorescent protein in mice. Microb. Cell Fact. 2015, 14, 181. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.C.; da Silva, A.C.; Oliveira, R.A.; Pereira, V.R.; Gil, L.H. In vivo near-infrared fluorescence imaging of leishmania amazonensis expressing infrared fluorescence protein (irfp) for real-time monitoring of cutaneous leishmaniasis in mice. J. Microbiol. Methods 2016, 130, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Graves, E.E.; Ripoll, J.; Weissleder, R.; Ntziachristos, V. A submillimeter resolution fluorescence molecular imaging system for small animal imaging. Med. Phys. 2003, 30, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.L.; Shcherbakova, D.M.; Verkhusha, V.V.; Kumar, A.T.N. In vivo tomographic imaging of deep-seated cancer using fluorescence lifetime contrast. Cancer Res. 2015, 75, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
| Bacterial Photoreceptor | Host | Size (Amino Acid Residues) | Chromophore | Absorbance (nm) | Em (nm) | Quantum Yield, % | Reference |
|---|---|---|---|---|---|---|---|
| Anacy 2551g3 | Anabaena cylindrica | 195 | PCB | 716, 730 | 740 | 1.2 | [58] |
| Cyan7822_4053g2 | Cyanothece sp. | 192 | PCB | 714, 732 | 736 | <1.0 | [58] |
| AM1_1557g2 | Acaryochloris marina | 165 | PCB | 649 | 676 | 1.7 | [46] |
| AM1_1557g2 | Acaryochloris marina | 165 | BV | 700 | 730 | 0.3 | [46] |
| AM1_C0023g2 | Acaryochloris marina | 151 | PCB | 650 | 879 | 3.0 | [46] |
| AM1_C0023g2 | Acaryochloris marina | 151 | BV | 699 | 718 | 0.2 | [46] |
| slr1393g3 | Synechocystis sp. | 162 | PCB | 650, 539 | 672, 616 | 6.0 | [83] |
| Npf2164g5 | Nostoc punctiforme | 170 | PCB | 662, 640 | n.a. | 10.0 | [75] |
| ApcE (36–240/Δ77–153) | Nostoc sp. | 130 | PCB | 650 | 663 | 6.0 | [88] |
| ApcD | Nostoc sp. | 160 | PCB | 650 | 663 | 7.4 | [89] |
| NIR FP | Ex, nm | Em, nm | EC, M−1·cm−1 | QY, % | Molecular Brightness vs. iRFP713, % | Oligomeric State | Photostability in Mammalian Cells, t1/2, s 1 | Brightness in HeLa Cells vs. iRFP713, % 2 | Reference |
|---|---|---|---|---|---|---|---|---|---|
| IFP1.4 | 684 | 708 | 92,000 | 7.7 | 114 | dimer | 70 | 8.0 | [32] |
| iRFP713 (aka iRFP) | 690 | 713 | 98,000 | 6.3 | 100 | dimer | 960 | 100 | [34] |
| PAiRFP1 | 659 3 | 703 3 | 67,100 | 4.8 | 64 | dimer | n.d. | 25 | [86] |
| PAiRFP2 | 692 3 | 719 3 | 63,600 | 4.7 | 60 | dimer | 25 | [86] | |
| IFP2.0 4 | 690 | 711 | 98,000 | 8.1 | 80 | dimer | 108 | 7.9 | [85] |
| iRFP670 | 643 | 670 | 114,000 | 12.2 | 225 | dimer | 290 | 119 | [10] |
| iRFP682 | 663 | 682 | 90,000 | 11.1 | 162 | dimer | 490 | 105 | [10] |
| iRFP702 | 673 | 702 | 93,000 | 8.2 | 124 | dimer | 630 | 61 | [10] |
| iRFP720 | 702 | 720 | 96,000 | 6.0 | 93 | dimer | 490 | 110 | [10] |
| smURFP | 642 | 670 | 180,000 | 18 | 551 | dimer | 300 | 1.0 | [101] |
| BphP1-FP | 640 | 669 | 60,000 | 13.0 | 126 | monomer | n.d. | n.d. | [27] |
| GAF-FP | 635 | 670 | 49,800 | 7.3 | 59 | monomer | n.d. | 2.0 | [12] |
| mIFP | 683 | 704 | 82,000 | 8.4 | 74 | monomer | 54 | 14 | [36] |
| Wi-Phy2 | 696 | 719 | 118 | 8,7 | n.d. | monomer | n.d. | n.d | [106] |
| miRFP670 | 642 | 670 | 87,400 | 14.0 | 198 | monomer | 155 | 72 | [35] |
| miRFP73 | 674 | 703 | 90,900 | 8.6 | 127 | monomer | 394 | 37 | [35] |
| miRFP79 | 683 | 709 | 78,400 | 5.4 | 69 | monomer | 192 | 30 | [35] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliinyk, O.S.; Chernov, K.G.; Verkhusha, V.V. Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes. Int. J. Mol. Sci. 2017, 18, 1691. https://doi.org/10.3390/ijms18081691
Oliinyk OS, Chernov KG, Verkhusha VV. Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes. International Journal of Molecular Sciences. 2017; 18(8):1691. https://doi.org/10.3390/ijms18081691
Chicago/Turabian StyleOliinyk, Olena S., Konstantin G. Chernov, and Vladislav V. Verkhusha. 2017. "Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes" International Journal of Molecular Sciences 18, no. 8: 1691. https://doi.org/10.3390/ijms18081691





