Clinical Applications of Autophagy Proteins in Cancer: From Potential Targets to Biomarkers
Abstract
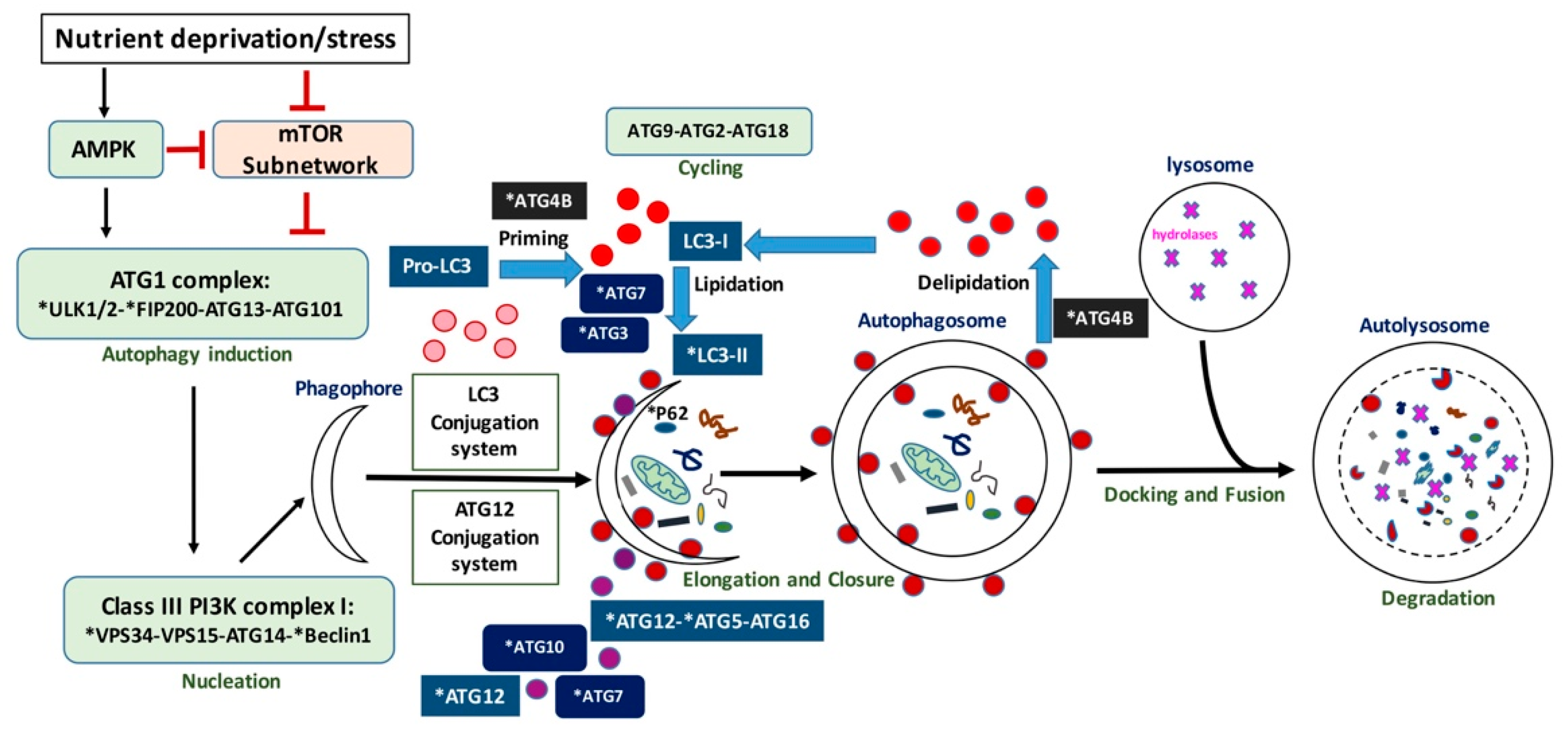
:1. Introduction
2. Autophagy Machinery
3. Candidate Autophagy-Related Biomarkers
3.1. LC3B
3.2. Other LC3/GABARAP Family Members
3.3. p62/SQSTM1
3.4. ULK-1/2
3.5. Beclin-1 and VPS34
3.6. ATG4B
3.7. Additional Autophagy-Related Biomarkers
3.8. Evaluation of an “Autophagy Signature” in Cancer
3.9. Secreted Factors as Potential Predictive Biomarkers
4. Discussion and Future Directions
Acknowledgments
Conflicts of Interest
References
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef] [PubMed]
- Rebecca, V.W.; Amaravadi, R.K. Emerging strategies to effectively target autophagy in cancer. Oncogene 2016, 35, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Solitro, A.R.; MacKeigan, J.P. Leaving the lysosome behind: Novel developments in autophagy inhibition. Future Med. Chem. 2016, 8, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, R.; Leone, R.; Chang, Y.C.; Fecher, L.A.; Schuchter, L.M.; Kramer, A.; Tan, K.-S.; Heitjan, D.F.; Rodgers, G.; Gallagher, M.; et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 2014, 10, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Poklepovic, A.; Gewirtz, D.A. Outcome of early clinical trials of the combination of hydroxychloroquine with chemotherapy in cancer. Autophagy 2014, 10, 1478–1480. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.R.; Ye, X.; Supko, J.G.; Desideri, S.; Grossman, S.A.; Brem, S.; Mikkelson, T.; Wang, D.; Chang, Y.C.; Hu, J.; et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014, 10, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Egan, D.F.; Chun, M.G.H.; Vamos, M.; Zou, H.; Rong, J.; Miller, C.J.; Lou, H.J.; Raveendra-Panickar, D.; Yang, C.-C.; Sheffler, D.J.; et al. Small molecule inhibition of the autophagy kinase ULK1 and Identification of ULK1 substrates. Mol. Cell 2015, 59, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Petherick, K.J.; Conway, O.J.L.; Mpamhanga, C.; Osborne, S.A.; Kamal, A.; Saxty, B.; Ganley, I.G. Pharmacological inhibition of ULK1 kinase blocks mammalian Target of Rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 2015, 290, 11376–11383. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.G.; Honson, N.S.; Arns, S.; Davis, T.L.; Dhe-Paganon, S.; Kovacic, S.; Kumar, N.S.; Pfeifer, T.A.; Young, R.N. Development of fluorescent substrates and assays for the key autophagy-related cysteine protease enzyme, ATG4B. Assay Drug Dev. Technol. 2014, 12, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Akin, D.; Wang, S.K.; Habibzadegah-Tari, P.; Law, B.; Ostrov, D.; Li, M.; Yin, X.-M.; Kim, J.-S.; Horenstein, N.; Dunn, W.A. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 2014, 10, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Bago, R.; Malik, N.; Munson, M.J.; Prescott, A.R.; Davies, P.; Sommer, E.; Shpiro, N.; Ward, R.; Cross, D.; Ganley, I.G.; et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014, 463, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, B.; El-Ahmad, Y.; Filoche-Rommé, B.; Dureuil, C.; Fassy, F.; Abecassis, P.-Y.; Mathieu, M.; Bertrand, T.; Benard, T.; Barrière, C.; et al. Discovery of (2S)-8-[(3R)-3-methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: A novel potent and selective inhibitor of Vps34 for the treatment of solid tumors. J. Med. Chem. 2015, 58, 376–400. [Google Scholar] [CrossRef] [PubMed]
- Dowdle, W.E.; Nyfeler, B.; Nagel, J.; Elling, R.A.; Liu, S.; Triantafellow, E.; Menon, S.; Wang, Z.; Honda, A.; Pardee, G.; et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014, 16, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.-F.; Lamberton, A.; Mathieu, M.; Bertrand, T.; et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014, 10, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Subramani, S.; Malhotra, V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013, 14, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A. Prognostic or predictive? It’s time to get back to definitions! J. Clin. Oncol. 2011, 29, 4718–4719. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Shelling, A.; Muthukaruppan, A.; Lasham, A.; Blenkiron, C.; Laking, G.; Print, C. Predictive and prognostic molecular markers for cancer medicine. Ther. Adv. Med. Oncol. 2010, 2, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Oldenhuis, C.N.A.M.; Oosting, S.F.; Gietema, J.A.; de Vries, E.G.E. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.-Y.; Xu, R.-H. Predictive and prognostic biomarkers with therapeutic targets in advanced colorectal cancer. World J. Gastroenterol. WJG 2014, 20, 3858–3874. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Hausdorf, D.; Schalper, K.A.; Neumeister, V.; Rimm, D.L. Quantitative measurement of cancer tissue biomarkers in the lab and in the clinic. Lab. Investig. 2015, 95, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Lazova, R.; Camp, R.L.; Klump, V.; Siddiqui, S.F.; Amaravadi, R.K.; Pawelek, J.M. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2012, 18, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Sarkar, S.; Davies, J.E.; Futter, M.; Garcia-Arencibia, M.; Green-Thompson, Z.W.; Jimenez-Sanchez, M.; Korolchuk, V.I.; Lichtenberg, M.; Luo, S.; et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010, 90, 1383–1435. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.K.; Lamming, D.W. The mechanistic target of rapamycin: The grand conducTOR of metabolism and aging. Cell Metab. 2016, 23, 990–1003. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: Autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 2013, 41, 1103–1130. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, F.; Williamson, L.E.; Tooze, S.A.; Chan, E.Y.W. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy 2013, 9, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian autophagy: How does it work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Levine, B. To be or not to be?: How selective autophagy and cell death govern cell fate. Cell 2014, 157, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Lebovitz, C.B.; Bortnik, S.B.; Gorski, S.M. Here, there be dragons: Charting autophagy-related alterations in human tumors. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2012, 18, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. “Protein modifications: Beyond the usual suspects” review series. EMBO Rep. 2008, 9, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Shvets, E.; Shpilka, T.; Shimron, F.; Shinder, V.; Elazar, Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010, 29, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Sou, Y.; Ezaki, J.; Minematsu-Ikeguchi, N.; Ueno, T.; Kominami, E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3-and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004, 279, 36268–36276. [Google Scholar] [CrossRef] [PubMed]
- Satoo, K.; Noda, N.N.; Kumeta, H.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009, 28, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, S.G.; Bakula, D.; Frickey, T.; Cezanne, A.; Brigger, D.; Tschan, M.P.; Robenek, H.; Proikas-Cezanne, T. Lipid droplet and early autophagosomal membrane targeting of Atg2A and Atg14L in human tumor cells. J. Lipid Res. 2014, 55, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A.; Thamm, D.H.; Gustafson, D.L. Autophagy and cancer therapy. Mol. Pharmacol. 2014, 85, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, J.; Ouyang, L.; Liu, B.; Cheng, Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett. 2016, 373, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.E.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M.A. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- McAfee, Q.; Zhang, Z.; Samanta, A.; Levi, S.M.; Ma, X.-H.; Piao, S.; Lynch, J.P.; Uehara, T.; Sepulveda, A.R.; Davis, L.E.; et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. USA 2012, 109, 8253–8258. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-H.; Piao, S.-F.; Dey, S.; Mcafee, Q.; Karakousis, G.; Villanueva, J.; Hart, L.S.; Levi, S.; Hu, J.; Zhang, G.; et al. Targeting ER stress–induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Investig. 2014, 124, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhao, X.; Subahan, N.R.; Fan, L.; Gao, J.; Chen, H. The prognostic value of autophagy-related markers beclin-1 and microtubule-associated protein light chain 3B in cancers: A systematic review and meta-analysis. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 7317–7326. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, M.; Zhao, J.; Wang, J.; Zhang, Y.; Zhang, Q. High expression of LC3B is associated with progression and poor outcome in triple-negative breast cancer. Med. Oncol. Northwood Lond. Engl. 2013, 30, 475. [Google Scholar] [CrossRef] [PubMed]
- Masuda, G.; Yashiro, M.; Kitayama, K.; Miki, Y.; Kasashima, H.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; Sakurai, K.; et al. Clinicopathological correlations of autophagy-related proteins LC3, Beclin 1 and p62 in gastric cancer. Anticancer Res. 2016, 36, 129–136. [Google Scholar] [PubMed]
- Cao, Q.-H.; Liu, F.; Yang, Z.-L.; Fu, X.-H.; Yang, Z.-H.; Liu, Q.; Wang, L.; Wan, X.-B.; Fan, X.-J. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am. J. Transl. Res. 2016, 8, 3831–3847. [Google Scholar] [PubMed]
- Zhou, H.; Yuan, M.; Yu, Q.; Zhou, X.; Min, W.; Gao, D. Autophagy regulation and its role in gastric cancer and colorectal cancer. Cancer Biomark. Sect. Dis. Markers 2016, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.J.; Ademi, C.; Bertram, S.; Schmid, K.W.; Baba, H.A. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J. Surg. Oncol. 2016, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhao, H.; Guo, L.; Zhang, Q.; Zhao, L.; Bai, S.; Zhang, M.; Xu, S.; Wang, F.; Wang, X.; et al. Autophagy-based survival prognosis in human colorectal carcinoma. Oncotarget 2015, 6, 7084–7103. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.-F.; Jiang, W.-Q.; Zhang, B.; Cai, Y.-C.; Xu, R.-H.; Chen, X.-X.; Wang, F.; Xia, L.-P. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J. Gastroenterol. 2011, 17, 4779–4786. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Sun, C.; Tian, D.; Li, Y.; Gao, X.; He, S.; Li, T. Expression and clinical significances of Beclin1, LC3 and mTOR in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 3882–3891. [Google Scholar] [PubMed]
- Winardi, D.; Tsai, H.-P.; Chai, C.-Y.; Chung, C.-L.; Loh, J.-K.; Chen, Y.-H.; Hsieh, C.-L. Correlation of altered expression of the autophagy marker lc3b with poor prognosis in astrocytoma. Bio. Med. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- El-Mashed, S.; O’Donovan, T.R.; Kay, E.W.; Abdallah, A.R.; Cathcart, M.-C.; O’Sullivan, J.; O’Grady, A.; Reynolds, J.; O’Reilly, S.; O’Sullivan, G.C.; et al. LC3B globular structures correlate with survival in esophageal adenocarcinoma. BMC Cancer 2015, 15, 582. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-L.; Chen, F.-F.; Lung, J.; Lo, C.-H.; Lee, F.-H.; Lu, Y.-C.; Hung, C.-H. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br. J. Cancer 2014, 111, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-H.; Jia, C.-C.; Chen, J.; Lin, Z.-X.; Ruan, D.-Y.; Li, X.; Lin, Q.; Dong, M.; Ma, X.-K.; Wan, X.-B.; et al. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 12225–12233. [Google Scholar] [CrossRef] [PubMed]
- Schläfli, A.M.; Adams, O.; Galván, J.A.; Gugger, M.; Savic, S.; Bubendorf, L.; Schmid, R.A.; Becker, K.-F.; Tschan, M.P.; Langer, R.; et al. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget 2016, 7, 39544–39555. [Google Scholar] [CrossRef] [PubMed]
- Ladoire, S.; Penault-Llorca, F.; Senovilla, L.; Dalban, C.; Enot, D.; Locher, C.; Prada, N.; Poirier-Colame, V.; Chaba, K.; Arnould, L.; et al. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy 2015, 11, 1878–1890. [Google Scholar] [CrossRef] [PubMed]
- Giatromanolaki, A.; Koukourakis, M.I.; Harris, A.L.; Polychronidis, A.; Gatter, K.C.; Sivridis, E. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J. Clin. Pathol. 2010, 63, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Sivridis, E.; Koukourakis, M.I.; Zois, C.E.; Ledaki, I.; Ferguson, D.J.P.; Harris, A.L.; Gatter, K.C.; Giatromanolaki, A. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am. J. Pathol. 2010, 176, 2477–2489. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Sun, L.; Wang, C.; Huang, H.; Liu, J.; Liao, W.; Shi, M. LC3A-positive “stonelike” structures predict an adverse prognosis of gastric cancer. Anat. Rec. 2014, 297, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Karpathiou, G.; Sivridis, E.; Koukourakis, M.I.; Mikroulis, D.; Bouros, D.; Froudarakis, M.E.; Giatromanolaki, A. Light-chain 3A autophagic activity and prognostic significance in non-small cell lung carcinomas. Chest 2011, 140, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Xi, S.-Y.; Lu, J.-B.; Chen, J.-W.; Cao, Y.; Luo, R.-Z.; Wu, Q.-L.; Cai, M.-Y. The “stone-like” pattern of LC3A expression and its clinicopathologic significance in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2013, 431, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Spowart, J.E.; Townsend, K.N.; Huwait, H.; Eshragh, S.; West, N.R.; Ries, J.N.; Kalloger, S.; Anglesio, M.; Gorski, S.M.; Watson, P.H.; et al. The autophagy protein LC3A correlates with hypoxia and is a prognostic marker of patient survival in clear cell ovarian cancer. J. Pathol. 2012, 228, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zhang, Y.; Chen, Y.; Chen, L.; Wang, F. GABARAP is overexpressed in colorectal carcinoma and correlates with shortened patient survival. Hepato-Gastroenterology 2010, 57, 257–261. [Google Scholar] [PubMed]
- Iwadate, R.; Inoue, J.; Tsuda, H.; Takano, M.; Furuya, K.; Hirasawa, A.; Aoki, D.; Inazawa, J. High expression of p62 protein is associated with poor prognosis and aggressive phenotypes in endometrial cancer. Am. J. Pathol. 2015, 185, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Iwadate, R.; Inoue, J.; Tsuda, H.; Takano, M.; Furuya, K.; Hirasawa, A.; Aoki, D.; Inazawa, J. High expression of SQSTM1/p62 protein is associated with poor prognosis in epithelial ovarian cancer. Acta. Histochem. Cytochem. 2014, 47, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Du, Z.; Li, L.; Shi, M.; Yu, Y. Beclin 1 and p62 expression in non-small cell lung cancer: Relation with malignant behaviors and clinical outcome. Int. J. Clin. Exp. Pathol. 2015, 8, 10644–10652. [Google Scholar] [PubMed]
- Inoue, D.; Suzuki, T.; Mitsuishi, Y.; Miki, Y.; Suzuki, S.; Sugawara, S.; Watanabe, M.; Sakurada, A.; Endo, C.; Uruno, A.; et al. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 2012, 103, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Rolland, P.; Madjd, Z.; Durrant, L.; Ellis, I.O.; Layfield, R.; Spendlove, I. The ubiquitin-binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr. Relat. Cancer 2007, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.-Z.; Yuan, Z.-Y.; Li, M.; Xi, S.-Y.; Fu, J.; He, J. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. OncoTargets Ther. 2013, 6, 883–888. [Google Scholar] [CrossRef]
- Zou, Y.; Chen, Z.; He, X.; He, X.; Wu, X.; Chen, Y.; Wu, X.; Wang, J.; Lan, P. High expression levels of unc-51-like kinase 1 as a predictor of poor prognosis in colorectal cancer. Oncol. Lett. 2015, 10, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Duan, B.-S.; Huang, J.-X.; Jiao, X.; Zhu, X.-W.; Sheng, H.-H.; Gao, H.-J.; Yu, H. Association of the expression of unc-51-Like kinase 1 with lymph node metastasis and survival in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Med. 2014, 7, 1349–1354. [Google Scholar] [PubMed]
- Xu, H.; Yu, H.; Zhang, X.; Shen, X.; Zhang, K.; Sheng, H.; Dai, S.; Gao, H. UNC51-like kinase 1 as a potential prognostic biomarker for hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 711–717. [Google Scholar] [PubMed]
- Yun, M.; Bai, H.-Y.; Zhang, J.-X.; Rong, J.; Weng, H.-W.; Zheng, Z.-S.; Xu, Y.; Tong, Z.-T.; Huang, X.-X.; Liao, Y.-J.; et al. ULK1: A promising biomarker in predicting poor prognosis and therapeutic response in human nasopharygeal carcinoma. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Deng, R.; Luo, R.-Z.; Shen, G.-P.; Cai, M.-Y.; Du, Z.-M.; Jiang, S.; Yang, M.-T.; Fu, J.-H.; Zhu, X.-F. Low expression of ULK1 is associated with operable breast cancer progression and is an adverse prognostic marker of survival for patients. Breast Cancer Res Treat. 2012, 134, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xue, X.-F.; Shen, H.-G.; Guo, X.-B.; Wang, X.; Yuan, B.; Guo, X.-P.; Kuang, Y.-T.; Zhi, Q.-M.; Zhao, H. Prognostic significance of Beclin-1 expression in colorectal cancer: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 4583–4587. [Google Scholar] [CrossRef] [PubMed]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Pitiakoudis, M.; Gatter, K.C.; Harris, A.L. Beclin 1 over-and underexpression in colorectal cancer: Distinct patterns relate to prognosis and tumour hypoxia. Br. J. Cancer 2010, 103, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-Y.; Gou, W.-F.; Zhao, S.; Mao, X.-Y.; Zheng, Z.-H.; Takano, Y.; Zheng, H.-C. Beclin 1 Expression is Closely Linked to Colorectal Carcinogenesis and Distant Metastasis of Colorectal Carcinoma. Int. J. Mol. Sci. 2014, 15, 14372–14385. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.K.; Kim, S.C.; Park, I.J.; Park, S.J.; Jin, D.-H.; Hong, S.-W.; Cho, D.-H.; Kim, J.C. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, L.; Zheng, Y.-Z.; Xu, Q.; Liu, X.-P.; Hu, X.; Wang, P.; Shao, Z.-M. Expression of autophagy-related proteins ATG5 and FIP200 predicts favorable disease-free survival in patients with breast cancer. Biochem. Biophys. Res. Commun. 2015, 458, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.H.; Cho, Y.-S.; Won, H.S.; Jeon, E.K.; An, H.J.; Hong, S.U.; Park, J.H.; Lee, M.A. Prognostic significance of autophagy-related protein expression in resected pancreatic ductal adenocarcinoma. Pancreas 2013, 42, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yamamoto, A.; Matsui, M.; Yoshimori, T.; Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 2004, 15, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, W.; Koo, J.S. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology 2013, 62, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.J.; Kim, Y.H.; Cho, N.H.; Koo, J.S. Expression of autophagy related proteins in invasive lobular carcinoma: Comparison to invasive ductal carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3389–3398. [Google Scholar] [PubMed]
- Guo, J.Y.; Chen, H.-Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.S.; Karantza, V.; et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes. Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; Xia, B.; White, E. Autophagy-mediated tumor promotion. Cell 2013, 155, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent insights into the function of autophagy in cancer. Genes. Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef] [PubMed]
- Puissant, A.; Fenouille, N.; Auberger, P. When autophagy meets cancer through p62/SQSTM1. Am. J. Cancer Res. 2012, 2, 397–413. [Google Scholar] [PubMed]
- Jiang, X.; Overholtzer, M.; Thompson, C.B. Autophagy in cellular metabolism and cancer. J. Clin. Investig. 2015, 125, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.G.; Hurley, J.H. Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 2016, 39, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, M.B.; Shokat, K.M. Discovery and structure of a new inhibitor scaffold of the autophagy initiating kinase ULK1. Bioorg. Med. Chem. 2015, 23, 5483–5488. [Google Scholar] [CrossRef] [PubMed]
- John Clotaire, D.Z.; Zhang, B.; Wei, N.; Gao, R.; Zhao, F.; Wang, Y.; Lei, M.; Huang, W. miR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem. Biophys. Res. Commun. 2016, 472, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Klionsky, D.J. Physiological functions of Atg6/Beclin 1: A unique autophagy-related protein. Cell Res. 2007, 17, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Kepp, O.; Michaud, M.; Martins, I.; Minoux, H.; Métivier, D.; Maiuri, M.C.; Kroemer, R.T.; Kroemer, G. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene 2011, 30, 4544–4556. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-F.; Leung, C.-M.; Chang, Y.-H.; Cheng, J.-S.; Chen, J.-J.; Weng, C.-J.; Tsai, K.-W.; Hsu, C.-J.; Liu, Y.-C.; Hsu, P.-C.; et al. ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy 2014, 10, 1454–1465. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Lin, H.; Lin, K.B.L.; Leung, A.; Wang, H.M.; Malekesmaeili, M.; Brinkman, R.R.; Forrest, D.L.; Gorski, S.M.; Jiang, X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 2014, 123, 3622–3634. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Su, J.; Qian, H.; Guo, T. SLC27A4 regulate ATG4B activity and control reactions to chemotherapeutics-induced autophagy in human lung cancer cells. Tumour. Biol. 2016, 37, 6943–6952. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, Z.; Tang, F.; Hu, P.; Yang, Z.; Xue, C.; Gong, J.; Shi, L.; Xie, C. ATG7 promotes the tumorigenesis of lung cancer but might be dispensable for prognosis predication: A clinicopathologic study. OncoTargets Ther. 2016, 9, 4975–4981. [Google Scholar] [CrossRef]
- Kim, M.S.; Song, S.Y.; Lee, J.Y.; Yoo, N.J.; Lee, S.H. Expressional and mutational analyses of ATG5 gene in prostate cancers. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2011, 119, 802–807. [Google Scholar] [CrossRef] [PubMed]
- An, C.H.; Kim, M.S.; Yoo, N.J.; Park, S.W.; Lee, S.H. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol. Res. Pract. 2011, 207, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-Y.; Fang, Y.-Y.; His, E.; Huang, Y.-C.; Hsu, N.C.-H.; Yang, W.-C.; Chang, H.-W.; Chai, C.-Y.; Chu, P.-Y. Immunopositivity of Beclin-1 and ATG5 as indicators of survival and disease recurrence in oral squamous cell carcinoma. Anticancer Res. 2013, 33, 5611–5616. [Google Scholar] [PubMed]
- Manjithaya, R.; Subramani, S. Autophagy: A broad role in unconventional protein secretion? Trends Cell Biol. 2011, 21, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kraya, A.A.; Piao, S.; Xu, X.; Zhang, G.; Herlyn, M.; Gimotty, P.; Levine, B.; Amaravadi, R.K.; Speicher, D.W. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy 2015, 11, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, inflammation, and immunity: A troika governing cancer and its treatment. Cell 2016, 166, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, D.; Mita, M.; Sarantopoulos, J.; Wood, L.; Amaravadi, R.K.; Davis, L.E.; Mita, A.C.; Curiel, T.J.; Espitia, C.M.; Nawrocki, S.T.; et al. Combined autophagy and HDAC inhibition. Autophagy 2014, 10, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Wani, W.Y.; Boyer-Guittaut, M.; Dodson, M.; Chatham, J.; Darley-Usmar, V.; Zhang, J. Regulation of autophagy by protein post-translational modification. Lab. Investig. 2015, 95, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Crutchfield, C.A.; Thomas, S.N.; Sokoll, L.J.; Chan, D.W. Advances in mass spectrometry-based clinical biomarker discovery. Clin. Proteom. 2016, 13. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Awad, N.; Kotb, Y. Identification and validation of a novel autophagy gene expression signature for human bladder cancer patients. Tumour. Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Lebovitz, C.B.; Robertson, A.G.; Goya, R.; Jones, S.J.; Morin, R.D.; Marra, M.A.; Gorski, S.M. Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy 2015, 11, 1668–1687. [Google Scholar] [CrossRef] [PubMed]
| Candidate Marker * | Prognostic Value ** | |
|---|---|---|
| Poor | Favorable | |
| LC3B | BC [22,48,49], GC [50,51,52], CRC [53,54,55,56], melanoma [22], astrocytoma [57], esophageal cancer [58], oral SCC [59], and HCC [60] | NSCLC [61], BC [62] |
| LC3A | CRC [63], BC [64], GC [65], NSCLCs [66], HCC [67] and clear cell OC [68] (stone-like structures) | CRC [63] and BC [64] (juxtanuclear accumulation) |
| GABARAP | CRC patients [69] | |
| p62 | endometrial cancer, [70], oral SCC [59], epithelial OC [71], and NSCLC [72,73], BC [74,75] | |
| ULK1 | CRC [76], esophageal SCC [77], HCC [78], and nasopharygeal carcinoma [79] | BC [80], GC [51] |
| Beclin1 | CRC [48,81] ***; [56,82,83] | GC, BC, NSCLC, CRC, lymphoma [48] *** |
| ATG3 | GC [51] | |
| ATG10 | CRC [84] | GC [51] |
| FIP200 | BC [85] | |
| Autophagy “signature”: ULK1, Beclin 1, ATG3, and ATG10 | GC [51] | |
| Autophagy “signature”: LC3B, ATG5, Beclin 1, Ambra1 and Bif-1 | PDAC [86] | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bortnik, S.; Gorski, S.M. Clinical Applications of Autophagy Proteins in Cancer: From Potential Targets to Biomarkers. Int. J. Mol. Sci. 2017, 18, 1496. https://doi.org/10.3390/ijms18071496
Bortnik S, Gorski SM. Clinical Applications of Autophagy Proteins in Cancer: From Potential Targets to Biomarkers. International Journal of Molecular Sciences. 2017; 18(7):1496. https://doi.org/10.3390/ijms18071496
Chicago/Turabian StyleBortnik, Svetlana, and Sharon M. Gorski. 2017. "Clinical Applications of Autophagy Proteins in Cancer: From Potential Targets to Biomarkers" International Journal of Molecular Sciences 18, no. 7: 1496. https://doi.org/10.3390/ijms18071496




