Multifaceted Interpretation of Colon Cancer Stem Cells
Abstract
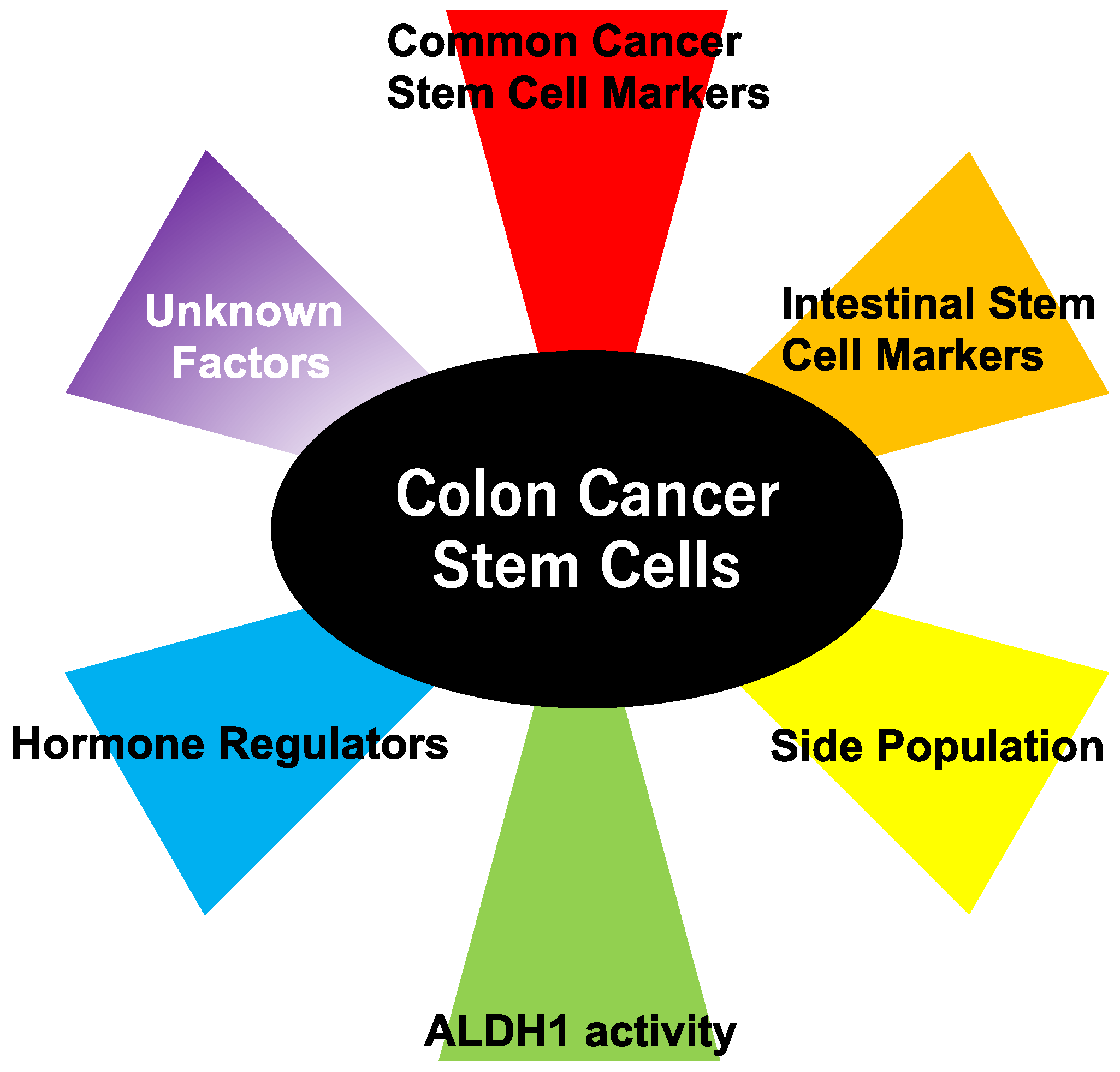
:1. Cancer Stem Cells (CSCs) and Colon Cancer
2. Methods to Detect Colon Cancer Stem Cells
2.1. Cell Surface CSC Markers
2.1.1. CD44
2.1.2. CD133
2.1.3. CD24
2.1.4. CD29
2.1.5. CD26
2.1.6. CD166
2.1.7. CD326
2.2. An Active Intestinal Stem Cell Marker, Leucine-Rich Repeat-Containing G-Protein-Coupled Receptor 5 (Lgr5)
2.3. Quiescent Intestinal Stem Cell Markers
2.4. CSC Markers of Migration
2.5. Side Population (SP) Cells
2.6. Aldehyde Dehydrogenase (ALDH) 1
2.7. Type III Deiodinase (D3)
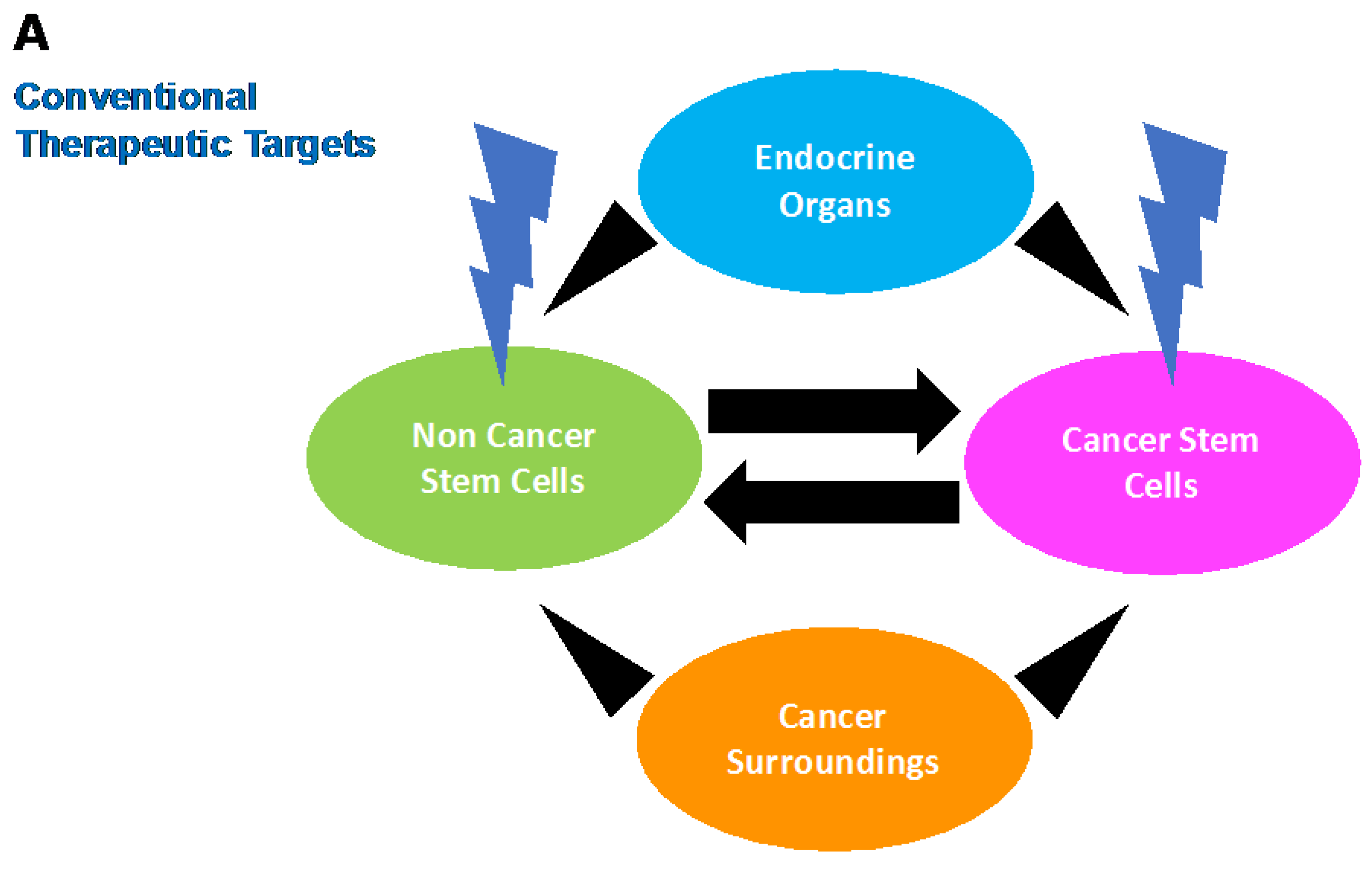
3. Stem Cell Phenotype in Colon Cancer: Emerging Evidence for Intertumoral Heterogeneity
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Wicha, M.S.; Liu, S.; Dontu, G. Cancer stem cells: An old idea—A paradigm shift. Cancer Res. 2006, 66, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; de Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Report 2014; International Agency for Research on Cancer (IARC): Lyon, France, 2014.
- Simmonds, P.C.; Primrose, J.N.; Colquitt, J.L.; Garden, O.J.; Poston, G.J.; Rees, M. Surgical resection of hepatic metastases from colorectal cancer: A systematic review of published studies. Br. J. Cancer 2006, 94, 982–999. [Google Scholar] [CrossRef] [PubMed]
- Tirino, V.; Desiderio, V.; Paino, F.; Papaccio, G.; de Rosa, M. Methods for cancer stem cell detection and isolation. Methods Mol. Biol. 2012, 879, 513–529. [Google Scholar] [PubMed]
- Yan, Y.; Zuo, X.; Wei, D. Concise review: Emerging role of CD44 in cancer stem cells: A promising biomarker and therapeutic target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Gunthert, U.; Hofmann, M.; Rudy, W.; Reber, S.; Zoller, M.; Haussmann, I.; Matzku, S.; Wenzel, A.; Ponta, H.; Herrlich, P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 1991, 65, 13–24. [Google Scholar] [CrossRef]
- Herrlich, P.; Pals, S.; Ponta, H. CD44 in colon cancer. Eur. J. Cancer 1995, 31A, 1110–1112. [Google Scholar] [CrossRef]
- Wielenga, V.J.; Heider, K.H.; Offerhaus, G.J.; Adolf, G.R.; van den Berg, F.M.; Ponta, H.; Herrlich, P.; Pals, S.T. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993, 53, 4754–4756. [Google Scholar] [PubMed]
- Mulder, J.W.; Kruyt, P.M.; Sewnath, M.; Oosting, J.; Seldenrijk, C.A.; Weidema, W.F.; Offerhaus, G.J.; Pals, S.T. Colorectal cancer prognosis and expression of exon-v6-containing CD44 proteins. Lancet 1994, 344, 1470–1472. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Chen, L.; Sleeman, J.P.; Herrlich, P.; Ponta, H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002, 16, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Tremmel, M.; Matzke, A.; Albrecht, I.; Laib, A.M.; Olaku, V.; Ballmer-Hofer, K.; Christofori, G.; Heroult, M.; Augustin, H.G.; Ponta, H.; et al. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood 2009, 114, 5236–5244. [Google Scholar] [CrossRef] [PubMed]
- Michieli, P.; Mazzone, M.; Basilico, C.; Cavassa, S.; Sottile, A.; Naldini, L.; Comoglio, P.M. Targeting the tumor and its microenvironment by a dual-function decoy met receptor. Cancer Cell 2004, 6, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Yaffe, M.B.; Sharp, P.A. A positive feedback loop couples ras activation and CD44 alternative splicing. Genes Dev. 2006, 20, 1715–1720. [Google Scholar] [CrossRef] [PubMed]
- Ohata, H.; Ishiguro, T.; Aihara, Y.; Sato, A.; Sakai, H.; Sekine, S.; Taniguchi, H.; Akasu, T.; Fujita, S.; Nakagama, H.; et al. Induction of the stem-like cell regulator CD44 by rho kinase inhibition contributes to the maintenance of colon cancer-initiating cells. Cancer Res. 2012, 72, 5101–5110. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. CD133: A stem cell biomarker and beyond. Exp. Hematol. Oncol. 2013, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; Todaro, M.; de Sousa Mello, F.; Sprick, M.R.; Kemper, K.; Perez Alea, M.; Richel, D.J.; Stassi, G.; Medema, J.P. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA 2008, 105, 13427–13432. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Gibson, P.; Currle, D.S.; Tong, Y.; Richardson, R.J.; Bayazitov, I.T.; Poppleton, H.; Zakharenko, S.; Ellison, D.W.; Gilbertson, R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 2009, 457, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; St Clair, R.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Sprick, M.R.; de Bree, M.; Scopelliti, A.; Vermeulen, L.; Hoek, M.; Zeilstra, J.; Pals, S.T.; Mehmet, H.; Stassi, G.; et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010, 70, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.Q.; Choi, Y.P.; Kang, S.; Youn, J.H.; Cho, N.H. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene 2010, 29, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Denkert, C.; Burkhardt, M.; Gansukh, T.; Bellach, J.; Altevogt, P.; Dietel, M.; Kristiansen, G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin. Cancer Res. 2005, 11, 6574–6581. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, H.W.; Hur, K.Y.; Kim, J.J.; Park, G.S.; Jang, S.H.; Song, Y.S.; Jang, K.S.; Paik, S.S. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J. Gastroenterol. 2009, 15, 2258–2264. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Fujimoto, K.; Beauchamp, R.D.; Whitehead, R.H. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 2002, 123, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.G.; Li, X.; Gray, P.D.; Kuang, J.; Clayton, F.; Samowitz, W.S.; Madison, B.B.; Gumucio, D.L.; Kuwada, S.K. Conditional deletion of β1 integrins in the intestinal epithelium causes a loss of hedgehog expression, intestinal hyperplasia, and early postnatal lethality. J. Cell Biol. 2006, 175, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Pietila, M.; Ivaska, J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br. J. Cancer 2016, 115, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Misumi, Y.; Hayashi, Y.; Arakawa, F.; Ikehara, Y. Molecular cloning and sequence analysis of human dipeptidyl peptidase iv, a serine proteinase on the cell surface. Biochim. Biophys. Acta 1992, 1131, 333–336. [Google Scholar] [CrossRef]
- Drucker, D.J.; Nauck, M.A. The incretin system: Glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 2006, 368, 1696–1705. [Google Scholar] [CrossRef]
- Pang, R.; Law, W.L.; Chu, A.C.; Poon, J.T.; Lam, C.S.; Chow, A.K.; Ng, L.; Cheung, L.W.; Lan, X.R.; Lan, H.Y.; et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell 2010, 6, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Yamochi, T.; Yamochi, T.; Aytac, U.; Ohnuma, K.; McKee, K.S.; Morimoto, C.; Dang, N.H. CD26 regulates p38 mitogen-activated protein kinase-dependent phosphorylation of integrin β1, adhesion to extracellular matrix, and tumorigenicity of T-anaplastic large cell lymphoma Karpas 299. Cancer Res. 2005, 65, 6950–6956. [Google Scholar] [CrossRef] [PubMed]
- de la Haba-Rodriguez, J.; Macho, A.; Calzado, M.A.; Blazquez, M.V.; Gomez, M.A.; Munoz, E.E.; Aranda, E. Soluble dipeptidyl peptidase IV (CD-26) in serum of patients with colorectal carcinoma. Neoplasma 2002, 49, 307–311. [Google Scholar] [PubMed]
- Lam, C.S.; Cheung, A.H.; Wong, S.K.; Wan, T.M.; Ng, L.; Chow, A.K.; Cheng, N.S.; Pak, R.C.; Li, H.S.; Man, J.H.; et al. Prognostic significance of CD26 in patients with colorectal cancer. PLoS ONE 2014, 9, e98582. [Google Scholar] [CrossRef] [PubMed]
- Lieto, E.; Galizia, G.; Orditura, M.; Romano, C.; Zamboli, A.; Castellano, P.; Mabilia, A.; Auricchio, A.; F, D.E.V.; Gemei, M. CD26-positive/CD326-negative circulating cancer cells as prognostic markers for colorectal cancer recurrence. Oncol. Lett. 2015, 9, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Bowen, M.A.; Patel, D.D.; Li, X.; Modrell, B.; Malacko, A.R.; Wang, W.C.; Marquardt, H.; Neubauer, M.; Pesando, J.M.; Francke, U.; et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 1995, 181, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Levin, T.G.; Powell, A.E.; Davies, P.S.; Silk, A.D.; Dismuke, A.D.; Anderson, E.C.; Swain, J.R.; Wong, M.H. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 2010, 139, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Zhang, Z.; Zhu, X.; Liu, Y.; Qu, D.; Wu, P.; Huang, J.; Xu, A.X. Prognostic value of CD166 expression in cancers of the digestive system: A systematic review and meta-analysis. PLoS ONE 2013, 8, e70958. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yang, W.; Zong, S.; Li, H.; Liu, S.; Li, W.; Shi, Q.; Hou, F. Clinicopathological, prognostic and predictive value of CD166 expression in colorectal cancer: A meta-analysis. Oncotarget 2017. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, S.V.; Velders, M.P.; Bakker, H.A.; Fleuren, G.J.; Warnaar, S.O. Ep-CAM: A human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 1994, 125, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.J.; Nagtegaal, I.D.; van Krieken, J.H.; Litvinov, S.V. The epithelial cell adhesion molecule (Ep-CAM) as a morphoregulatory molecule is a tool in surgical pathology. Am. J. Pathol. 2003, 163, 2139–2148. [Google Scholar] [CrossRef]
- Balzar, M.; Prins, F.A.; Bakker, H.A.; Fleuren, G.J.; Warnaar, S.O.; Litvinov, S.V. The structural analysis of adhesions mediated by Ep-CAM. Exp. Cell. Res. 1999, 246, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Salem, R.R.; Wolf, B.C.; Sears, H.F.; Lavin, P.T.; Ravikumar, T.S.; DeCoste, D.; D’Emilia, J.C.; Herlyn, M.; Schlom, J.; Gottlieb, L.S.; et al. Expression of colorectal carcinoma-associated antigens in colonic polyps. J. Surg. Res. 1993, 55, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Went, P.; Vasei, M.; Bubendorf, L.; Terracciano, L.; Tornillo, L.; Riede, U.; Kononen, J.; Simon, R.; Sauter, G.; Baeuerle, P.A. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br. J. Cancer 2006, 94, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Munz, M.; Baeuerle, P.A.; Gires, O. The emerging role of epcam in cancer and stem cell signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Budhu, A.; Forgues, M.; Wang, X.W. Activation of hepatic stem cell marker epcam by Wnt-β-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007, 67, 10831–10839. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am. J. Anat. 1974, 141, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors Lgr4 and Lgr5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.G.; Snippert, H.J.; Stange, D.E.; van den Born, M.; van Es, J.H.; van de Wetering, M.; Clevers, H. Lineage tracing reveals lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012, 337, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Preston, S.L.; Wong, W.M.; Chan, A.O.; Poulsom, R.; Jeffery, R.; Goodlad, R.A.; Mandir, N.; Elia, G.; Novelli, M.; Bodmer, W.F.; et al. Bottom-up histogenesis of colorectal adenomas: Origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res. 2003, 63, 3819–3825. [Google Scholar] [PubMed]
- Shih, I.M.; Wang, T.L.; Traverso, G.; Romans, K.; Hamilton, S.R.; Ben-Sasson, S.; Kinzler, K.W.; Vogelstein, B. Top-down morphogenesis of colorectal tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 2640–2645. [Google Scholar] [CrossRef] [PubMed]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Goktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Potten, C.S. Extreme sensitivity of some intestinal crypt cells to X and γ irradiation. Nature 1977, 269, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Buczacki, S.J.; Zecchini, H.I.; Nicholson, A.M.; Russell, R.; Vermeulen, L.; Kemp, R.; Winton, D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013, 495, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Haupt, Y.; Alexander, W.S.; Barri, G.; Klinken, S.P.; Adams, J.M. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in Eµ-myc transgenic mice. Cell 1991, 65, 753–763. [Google Scholar] [CrossRef]
- Lessard, J.; Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003, 423, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Oguro, H.; Yuan, J.; Ichikawa, H.; Ikawa, T.; Yamazaki, S.; Kawamoto, H.; Nakauchi, H.; Iwama, A. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 2010, 6, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Sangiorgi, E.; Capecchi, M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008, 40, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Biehs, B.; Warming, S.; Leong, K.G.; Rangell, L.; Klein, O.D.; de Sauvage, F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 2011, 478, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; van Galen, P.; Pedley, N.M.; Lima-Fernandes, E.; Frelin, C.; Davis, T.; Cao, L.; Baiazitov, R.; Du, W.; Sydorenko, N.; et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 2014, 20, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Ohta, M.; Kanai, F.; Guleng, B.; Tanaka, Y.; Asaoka, Y.; Tada, M.; Seto, M.; Jazag, A.; Lianjie, L.; et al. Dysregulated expression of stem cell factor bmi1 in precancerous lesions of the gastrointestinal tract. Clin. Cancer Res. 2006, 12, 6960–6966. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, Y.; Li, J.; Zheng, J. Polycomb group protein Bmi1 expression in colon cancers predicts the survival. Med. Oncol. 2010, 27, 1273–1276. [Google Scholar] [CrossRef] [PubMed]
- Li, D.W.; Tang, H.M.; Fan, J.W.; Yan, D.W.; Zhou, C.Z.; Li, S.X.; Wang, X.L.; Peng, Z.H. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Pun, J.C.; Chan, J.Y.; Chun, B.K.; Ng, K.W.; Tsui, S.Y.; Wan, T.M.; Lo, O.; Poon, J.T.; Ng, L.; Pang, R. Plasma Bmi1 mRNA as a potential prognostic biomarker for distant metastasis in colorectal cancer patients. Mol. Clin. Oncol. 2014, 2, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Katoh, H.; Watanabe, M. The homeobox only protein homeobox (HOPX) and colorectal cancer. Int. J. Mol. Sci. 2013, 14, 23231–23243. [Google Scholar] [CrossRef] [PubMed]
- May, R.; Riehl, T.E.; Hunt, C.; Sureban, S.M.; Anant, S.; Houchen, C.W. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 2008, 26, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.K.; Carlone, D.L.; Richmond, C.A.; Farilla, L.; Kranendonk, M.E.; Henderson, D.E.; Baffour-Awuah, N.Y.; Ambruzs, D.M.; Fogli, L.K.; Algra, S.; et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Wang, Y.; Li, Y.; Poulin, E.J.; Means, A.L.; Washington, M.K.; Higginbotham, J.N.; Juchheim, A.; Prasad, N.; Levy, S.E.; et al. The Pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012, 149, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Jung, A.; Spaderna, S.; Hlubek, F.; Kirchner, T. Opinion: Migrating cancer stem cells — An integrated concept of malignant tumour progression. Nat. Rev. Cancer 2005, 5, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Conacci-Sorrell, M.; Simcha, I.; Ben-Yedidia, T.; Blechman, J.; Savagner, P.; Ben-Ze’ev, A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: The roles of β-catenin signaling, slug, and mapk. J. Cell Biol. 2003, 163, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Bain, G.; Wang, X.; Papkoff, J. Regulation of epithelial cell migration and tumor formation by β-catenin signaling. Exp. Cell Res. 2002, 280, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Eger, A.; Stockinger, A.; Park, J.; Langkopf, E.; Mikula, M.; Gotzmann, J.; Mikulits, W.; Beug, H.; Foisner, R. β-catenin and TGFβ signalling cooperate to maintain a mesenchymal phenotype after foser-induced epithelial to mesenchymal transition. Oncogene 2004, 23, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Busch, E.L.; McGraw, K.A.; Sandler, R.S. The potential for markers of epithelial-mesenchymal transition to improve colorectal cancer outcomes: A systematic review. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Hirschmann-Jax, C.; Foster, A.E.; Wulf, G.G.; Nuchtern, J.G.; Jax, T.W.; Gobel, U.; Goodell, M.A.; Brenner, M.K. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 14228–14233. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, N.; Utsunomiya, T.; Inoue, H.; Tanaka, F.; Mimori, K.; Barnard, G.F.; Mori, M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells 2006, 24, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Ma, L.; Hu, X.; Zhang, C.; Cheng, Y. Characterization of side population cells isolated from the colon cancer cell line SW480. Int J. Oncol. 2014, 45, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M. The hypoxic cell: A target for selective cancer therapy—Eighteenth Bruce F. Cain memorial award lecture. Cancer Res. 1999, 59, 5863–5870. [Google Scholar] [PubMed]
- Fletcher, J.I.; Haber, M.; Henderson, M.J.; Norris, M.D. Abc transporters in cancer: More than just drug efflux pumps. Nat. Rev. Cancer 2010, 10, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Meijer, G.A.; Schroeijers, A.B.; Flens, M.J.; Meuwissen, S.G.; van der Valk, P.; Baak, J.P.; Scheper, R.J. Increased expression of multidrug resistance related proteins Pgp, MRP1, and LRP/MVP occurs early in colorectal carcinogenesis. J. Clin. Pathol. 1999, 52, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ocana, A.; Tannock, I.F. Reversal of ATP-binding cassette drug transporter activity to modulate chemoresistance: Why has it failed to provide clinical benefit? Cancer Metastasis Rev. 2013, 32, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [PubMed]
- Ordonez-Moran, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Dolle, L.; Boulter, L.; Leclercq, I.A.; van Grunsven, L.A. Next generation of aldh substrates and their potential to study maturational lineage biology in stem and progenitor cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G573–G578. [Google Scholar] [CrossRef] [PubMed]
- Sirakov, M.; Skah, S.; Nadjar, J.; Plateroti, M. Thyroid hormone’s action on progenitor/stem cell biology: New challenge for a classic hormone? Biochim. Biophys. Acta 2013, 1830, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Dentice, M.; Ambrosio, R.; Luongo, C.; Carollo, R.; Benfante, A.; Todaro, M.; Stassi, G.; Salvatore, D. Activated thyroid hormone promotes differentiation and chemotherapeutic sensitization of colorectal cancer stem cells by regulating Wnt and Bmp4 signaling. Cancer Res. 2016, 76, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- De Sousa, E.M.F.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Merlos-Suarez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Cespedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; da Silva-Diz, V.; Munoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, E.M.F.; Colak, S.; Buikhuisen, J.; Koster, J.; Cameron, K.; de Jong, J.H.; Tuynman, J.B.; Prasetyanti, P.R.; Fessler, E.; van den Bergh, S.P.; et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011, 9, 476–485. [Google Scholar]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.; Lannon, W.A.; Grotzinger, C.; del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Hatano, Y.; Semi, K.; Hashimoto, K.; Lee, M.S.; Hirata, A.; Tomita, H.; Kuno, T.; Takamatsu, M.; Aoki, K.; Taketo, M.M.; et al. Reducing DNA methylation suppresses colon carcinogenesis by inducing tumor cell differentiation. Carcinogenesis 2015, 36, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Semi, K.; Yamada, Y. Application of iPS cell technology to cancer epigenome study: Uncovering the mechanism of cell status conversion for drug resistance in tumor. Pathol. Int. 2014, 64, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Yamada, Y.; Semi, K.; Yagi, M.; Tanaka, A.; Itakura, F.; Aoki, H.; Kunisada, T.; Woltjen, K.; Haga, H.; et al. Cellular context-dependent consequences of Apc mutations on gene regulation and cellular behavior. Proc. Natl. Acad. Sci. USA 2017, 114, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; DeWitt, N. In vivo cellular reprogramming: The next generation. Cell 2016, 166, 1386–1396. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatano, Y.; Fukuda, S.; Hisamatsu, K.; Hirata, A.; Hara, A.; Tomita, H. Multifaceted Interpretation of Colon Cancer Stem Cells. Int. J. Mol. Sci. 2017, 18, 1446. https://doi.org/10.3390/ijms18071446
Hatano Y, Fukuda S, Hisamatsu K, Hirata A, Hara A, Tomita H. Multifaceted Interpretation of Colon Cancer Stem Cells. International Journal of Molecular Sciences. 2017; 18(7):1446. https://doi.org/10.3390/ijms18071446
Chicago/Turabian StyleHatano, Yuichiro, Shinya Fukuda, Kenji Hisamatsu, Akihiro Hirata, Akira Hara, and Hiroyuki Tomita. 2017. "Multifaceted Interpretation of Colon Cancer Stem Cells" International Journal of Molecular Sciences 18, no. 7: 1446. https://doi.org/10.3390/ijms18071446
APA StyleHatano, Y., Fukuda, S., Hisamatsu, K., Hirata, A., Hara, A., & Tomita, H. (2017). Multifaceted Interpretation of Colon Cancer Stem Cells. International Journal of Molecular Sciences, 18(7), 1446. https://doi.org/10.3390/ijms18071446




