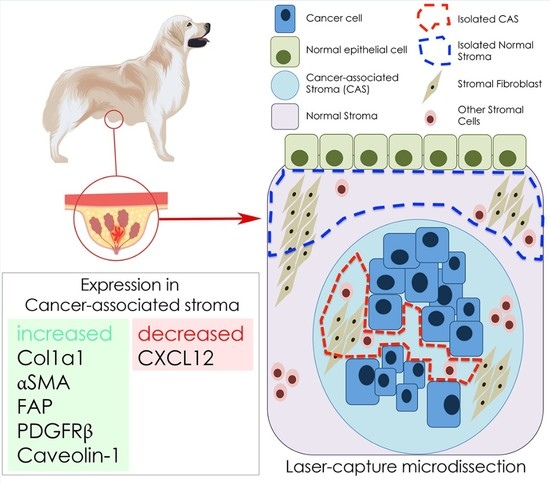
Analysis of Gene Expression Signatures in Cancer-Associated Stroma from Canine Mammary Tumours Reveals Molecular Homology to Human Breast Carcinomas
Abstract
:1. Introduction
2. Results
2.1. Selection of Cases Included in the Study
2.2. Selection of Cancer Associated Stroma Markers to be Analysed in This Study
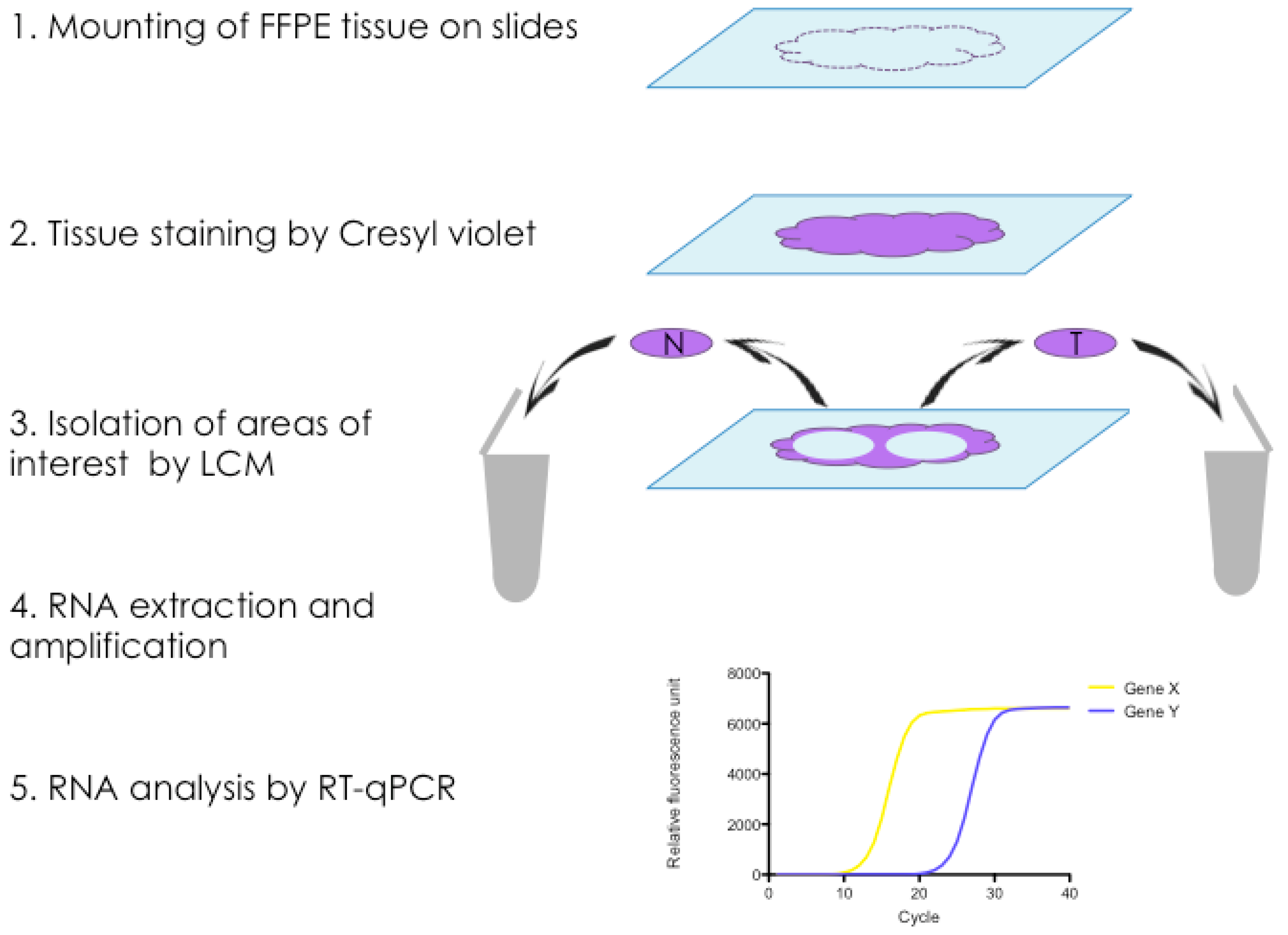
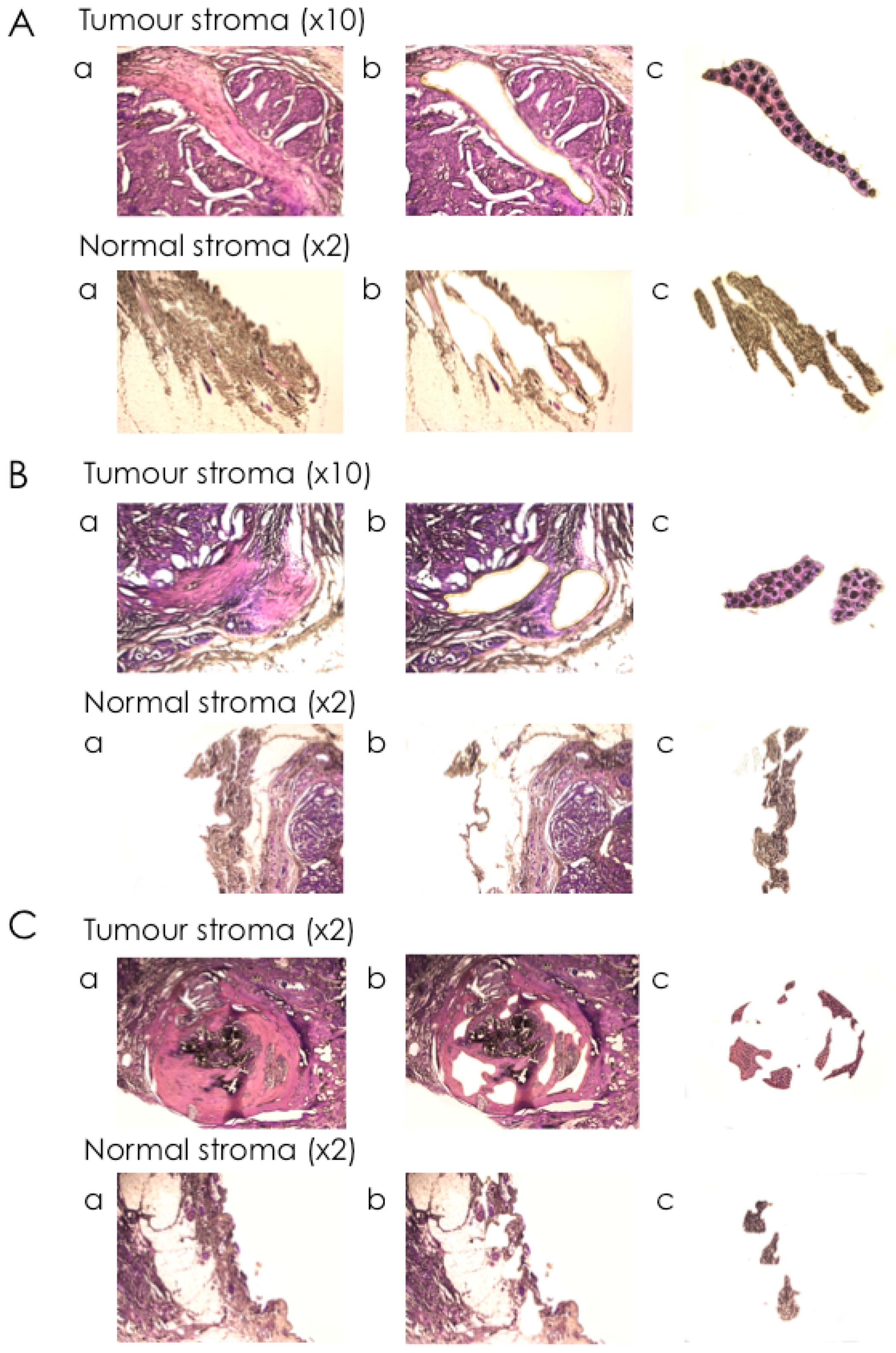
2.3. Isolation of mRNA from Tumour Stroma and Matched Normal Stroma from Patient Material
2.4. Expression Analysis of Cancer Associated Stroma Markers by RT-qPCR
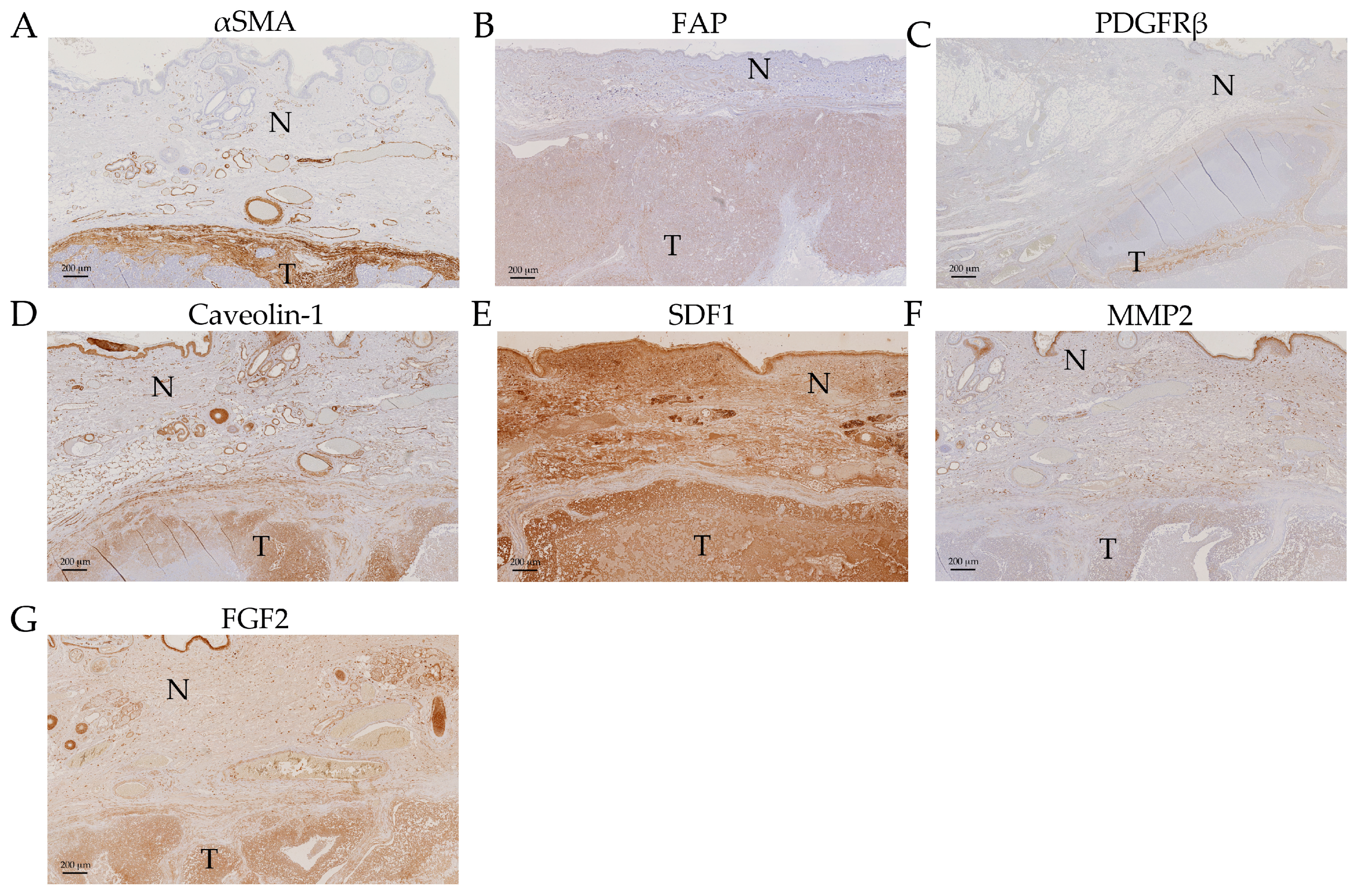
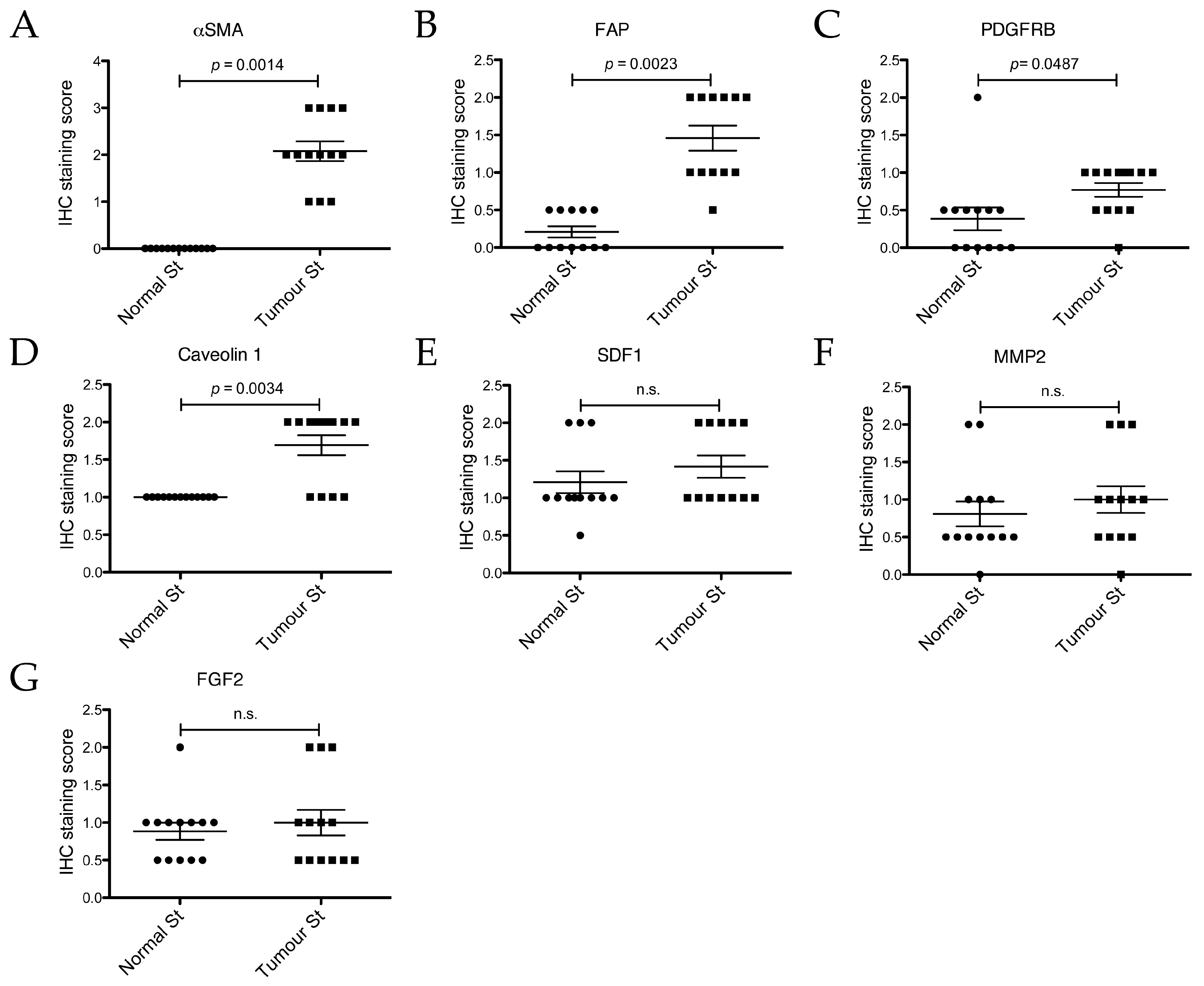
2.5. Expression Analysis of Cancer Associated Stroma Markers by Immunohistochemistry
3. Discussion
3.1. ACTA2/αSMA
3.2. COL1A1/Collagen 1
3.3. Fibroblast Activation Protein, FAP
3.4. PDGFRB/PDGFRβ
3.5. CXCL12/SDF1
3.6. MMP2
3.7. Interleukin 6, IL6
3.8. Caveolin-1
4. Materials and Methods
4.1. Selection of Cases for LCM
4.2. Tissue Processing and Staining
4.3. Laser-Capture Microdissection (LCM)
4.4. Isolation of mRNA from FFPE Tissue Sections Isolated by LCM
4.5. cDNA Synthesis and Preamplification
4.6. Reverse Transcription quantitative PCR
4.7. Graphical Display of Results and Statistical Analysis
4.8. Immunohistochemistry
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functionsof Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Tu, G.; Liu, Z.; Liu, M. Cancer-associated fibroblasts: A multifaceted driver of breast cancer progression. Cancer Lett. 2015, 361, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.J.; Radisky, D. Putting tumours in context. Nat. Rev. Cancer 2001, 1, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.F.; Iglesias, M.; Céspedes, M.V.; Sevillano, M.; Nadal, C.; Jung, P.; Zhang, X.; et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.F.; Byrom, D. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; Bertos, N.; Pepin, F.; Sadekova, S.; Souleimanova, M.; Zhao, H.; Chen, H.; Omeroglu, G.; Meterissian, S.; Omeroglu, A. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008, 14, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Pepin, F.; Bertos, N.; Laferriere, J.; Sadekova, S.; Souleimanova, M.; Zhao, H.; Finak, G.; Meterissian, S.; Hallett, M.T.; Park, M. Gene expression profiling of microdissected breast cancer microvasculature identifies distinct tumor vascular subtypes. Breast Cancer Res. 2012, 14, R120. [Google Scholar] [CrossRef] [PubMed]
- Rogers, N. Canine clues: Dog genomes explored in effort to bring human cancer to heel. Nat. Med. 2015, 21, 1374–1375. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.K.; Lindblad-Toh, K. Leader of the pack: Gene mapping in dogs and other model organisms. Nat. Rev. Genet. 2008, 9, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Gardner, H.L.; Fenger, J.M.; London, C.A. Dogs as a Model for Cancer. Annu. Rev. Anim. Biosci. 2015, 4, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xiong, H.; Ellis, A.E.; Northrup, N.C.; Rodriguez, C.O.; O’Regan, R.M.; Dalton, S.; Zhao, S. Molecular homology and difference between spontaneous canine mammary cancer and human breast cancer. Cancer Res. 2014, 74, 5045–5056. [Google Scholar] [CrossRef] [PubMed]
- Queiroga, F.L.; Raposo, T.; Carvalho, M.I.; Prada, J.; Pires, I. Canine mammary tumours as a model to study human breast cancer: Most recent findings. In Vivo 2011, 25, 455–465. [Google Scholar] [PubMed]
- Kessler, M. Kleintieronkologie. Diagnose und Therapie von Tumorerkrankungen bei. Hund und Katze, 3rd ed.; Enke Verlag: Stuttgart, Germany, 2012. [Google Scholar]
- Salas, Y.; Márquez, A.; Diaz, D.; Romero, L. Epidemiological Study of Mammary Tumors in Female Dogs Diagnosed during the Period 2002–2012: A Growing Animal Health Problem. PLoS ONE 2015, 10, e0127381. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, J.; Ehnman, M.; Ostman, A. PDGF receptors in tumor biology: Prognostic and predictive potential. Future Oncol. 2014, 10, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, J.; Sjöblom, T.; Micke, P.; Pontén, F.; Landberg, G.; Heldin, C.-H.; Bergh, J.; Brennan, D.J.; Jirström, K.; Ostman, A. Prognostic significance of stromal platelet-derived growth factor beta-receptor expression in human breast cancer. Am. J. Pathol. 2009, 175, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Frings, O.; Augsten, M.; Tobin, N.P.; Carlson, J.; Paulsson, J.; Pena, C.; Olsson, E.; Veerla, S.; Bergh, J.; Ostman, A.; et al. Prognostic significance in breast cancer of a gene signature capturing stromal PDGF signaling. Am. J. Pathol. 2013, 182, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A.; et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-J.; Dahiya, S.; Richardson, E.; Erlander, M.; Sgroi, D.C. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009, 11, R7. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, A.; Tagliabue, E.; Sørlie, T.; Naume, B.; Triulzi, T.; Orlandi, R.; Russnes, H.G.; Nesland, J.M.; Tammi, R.; Auvinen, P.; et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J. Pathol. 2008, 214, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Triulzi, T.; Casalini, P.; Sandri, M.; Ratti, M.; Carcangiu, M.L.; Colombo, M.P.; Balsari, A.; Ménard, S.; Orlandi, R.; Tagliabue, E. Neoplastic and stromal cells contribute to an extracellular matrix gene expression profile defining a breast cancer subtype likely to progress. PLoS ONE 2013, 8, e56761. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.H.; Espinosa, I.; Gilks, C.B.; van de Rijn, M.; West, R.B. The fibromatosis signature defines a robust stromal response in breast carcinoma. Lab. Investig. 2008, 88, 591–601. [Google Scholar] [CrossRef] [PubMed]
- West, R.B.; Nuyten, D.S.A.; Subramanian, S.; Nielsen, T.O.; Corless, C.L.; Rubin, B.P.; Montgomery, K.; Zhu, S.; Patel, R.; Hernandez-Boussard, T.; et al. Determination of stromal signatures in breast carcinoma. PLoS Biol. 2005, 3, e187. [Google Scholar] [CrossRef] [PubMed]
- Helleman, J.; Jansen, M.P.H.M.; Ruigrok-Ritstier, K.; van Staveren, I.L.; Look, M.P.; Meijer-van Gelder, M.E.; Sieuwerts, A.M.; Klijn, J.G.M.; Sleijfer, S.; et al. Association of an Extracellular Matrix Gene Cluster with Breast Cancer Prognosis and Endocrine Therapy Response. Clin. Cancer Res. 2008, 14, 5555–5564. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Tumor Immunotherapy Targeting Fibroblast Activation Protein, a Product Expressed in Tumor-Associated Fibroblasts. Cancer Res. 2005, 65, 11156–11163. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Lenter, M.C.; Zimmermann, R.N.; Garin-Chesa, P.; Old, L.J.; Rettig, W.J. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J. Biol. Chem. 1999, 274, 36505–36512. [Google Scholar] [CrossRef] [PubMed]
- Surowiak, P.; Murawa, D.; Materna, V.; Maciejczyk, A.; Pudelko, M.; Ciesla, S.; Breborowicz, J.; Murawa, P.; Zabel, M.; Dietel, M.; et al. Occurence of stromal myofibroblasts in the invasive ductal breast cancer tissue is an unfavourable prognostic factor. Anticancer Res. 2007, 27, 2917–2924. [Google Scholar] [PubMed]
- Sappino, A.P.; Skalli, O.; Jackson, B.; Schürch, W.; Gabbiani, G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int. J. Cancer 1988, 41, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Elenbaas, B.; Weinberg, R.A. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp. Cell Res. 2001, 264, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Ogawa, T.; Zhang, X.; Hanamura, N.; Kashikura, Y.; Takamura, M.; Yoneda, M.; Shiraishi, T. Role of stromal myofibroblasts in invasive breast cancer: Stromal expression of α-smooth muscle actin correlates with worse clinical outcome. Breast Cancer 2012, 19, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Surowiak, P.; Suchocki, S.; Győrffy, B.; Gansukh, T.; Wojnar, A.; Maciejczyk, A.; Pudełko, M.; Zabel, M. Stromal myofibroblasts in breast cancer: Relations between their occurrence, tumor grade and expression of some tumour markers. Folia Histochem. Cytobiol. 2006, 44, 111–116. [Google Scholar] [PubMed]
- Yazhou, C.; Wenlv, S.; Weidong, Z.; Licun, W. Clinicopathological significance of stromal myofibroblasts in invasive ductal carcinoma of the breast. Tumour Biol. 2004, 25, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Acar, A.; Eaton, E.N.; Mellody, K.T.; Scheel, C.; Ben-Porath, I.; Onder, T.T.; Wang, Z.C.; Richardson, A.L.; Weinberg, R.A.; et al. Autocrine TGF-β and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. USA 2010, 107, 20009–20014. [Google Scholar] [CrossRef] [PubMed]
- Chavey, C.; Bibeau, F.; Gourgou-Bourgade, S.; Burlinchon, S.; Boissière, F.; Laune, D.; Roques, S.; Lazennec, G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007, 9, R15. [Google Scholar] [CrossRef] [PubMed]
- Schafer, Z.T.; Brugge, J.S. IL-6 involvement in epithelial cancers. J. Clin. Investig. 2007, 117, 3660–3663. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; Dasgupta, A.; Sotgia, F.; Mercier, I.; Pestell, R.G.; Sabel, M.; Kleer, C.G.; Brody, J.R.; Lisanti, M.P. An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am. J. Pathol. 2009, 174, 2023–2034. [Google Scholar] [CrossRef] [PubMed]
- Burgemeister, R. Nucleic acids extraction from laser microdissected FFPE tissue sections. Methods Mol. Biol. 2011, 724, 117–129. [Google Scholar] [PubMed]
- Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Caveolae and signalling in cancer. Nat. Rev. Cancer 2015, 15, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Zischek, C. Das Tumorstroma als Angriffspunkt Einer Stammzellbasierten CCL5-Promoter/HSV-TK Suizidgentherapie in einem Murinen Pankreastumormodell. Ph.D. Thesis, Ludwig-Maximilians-Universität München, Munich, Germany, 2011. [Google Scholar]
- Gandellini, P.; Andriani, F.; Merlino, G.; D’Aiuto, F.; Roz, L.; Callari, M. Complexity in the tumour microenvironment: Cancer associated fibroblast gene expression patterns identify both common and unique features of tumour-stroma crosstalk across cancer types. Semin. Cancer Biol. 2015, 35. [Google Scholar] [CrossRef] [PubMed]
- Mihala, A.; Alexa, A.A.; Samoilă, C.; Dema, A.; Vizitiu, A.C.; Anghel, A.; Tămaş, L.; Marian, C.V.; Sîrbu, I. A pilot study on the expression of microRNAs resident on chromosome 21 in laser microdissected FFPE prostate adenocarcinoma samples. Rom. J. Morphol. Embryol. 2015, 56, 1063–1068. [Google Scholar] [PubMed]
- Morton, M.L.; Bai, X.; Merry, C.R.; Linden, P.A.; Khalil, A.M.; Leidner, R.S.; Thompson, C.L. Identification of mRNAs and lincRNAs associated with lung cancer progression using next-generation RNA sequencing from laser micro-dissected archival FFPE tissue specimens. Lung Cancer 2014, 85, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; Sadekova, S.; Pepin, F.; Hallett, M.; Meterissian, S.; Halwani, F.; Khetani, K.; Souleimanova, M.; Zabolotny, B.; Omeroglu, A.; et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006, 8, R58. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, H.; Michishita, M.; Ohkusu-Tsukada, K.; Takahashi, K. Increased presence of stromal myofibroblasts and tenascin-C with malignant progression in canine mammary tumors. Vet. Pathol. 2011, 48, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Giussani, M.; Merlino, G.; Cappelletti, V.; Tagliabue, E.; Daidone, M.G. Tumor-extracellular matrix interactions: Identification of tools associated with breast cancer progression. Semin. Cancer Biol. 2015, 35, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.; Santos Horta Dos, R.; Constantino-Casas, F.; Hoather, T.; Dobson, J. Expression of Fibroblast Activating Protein and Correlation with Histological Grade, Mitotic Index and Ki67 Expression in Canine Mast Cell Tumours. J. Comp. Pathol. 2017, 156, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J. Tumor stroma as targets for cancer therapy. Pharmacol. Ther. 2013, 137, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Lopes, C.; Frias, C.; Amorim, I.; Vicente, C.; Gärtner, F.; Matos, A. Immunohistochemical evaluation of MMP-2 and TIMP-2 in canine mammary tumours: A survival study. Vet. J. 2011, 190, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Aresu, L.; Giantin, M.; Morello, E.; Vascellari, M.; Castagnaro, M.; Lopparelli, R.; Zancanella, V.; Granato, A.; Garbisa, S.; Aricò, A.; et al. Matrix metalloproteinases and their inhibitors in canine mammary tumors. BMC Vet. Res. 2011, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-W.; Xu, K.-L.; Ruan, S.-Q.; Zhao, L.-L.; Chen, R.-L. Overexpression of caveolin-1 in cancer-associated fibroblasts predicts good outcome in breast cancer. Breast Care 2012, 7, 477–483. [Google Scholar] [PubMed]
- Shinoda, H.; Legare, M.E.; Mason, G.L.; Berkbigler, J.L.; Afzali, M.F.; Flint, A.F.; Hanneman, W.H. Significance of ERα, HER2, and CAV1 expression and molecular subtype classification to canine mammary gland tumor. J. Vet. Diagn. Investig. 2014, 26, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.G.; Minguet, S.; Navarro-Lérida, I.; Lazcano, J.J.; Samaniego, R.; Calvo, E.; Tello, M.; Osteso-Ibáñez, T.; Pellinen, T.; Echarri, A.; et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell 2011, 146, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Rowell, J.L.; McCarthy, D.O.; Alvarez, C.E. Dog models of naturally occurring cancer. Trends Mol. Med. 2011, 17, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Hester, S.D.; Bhat, V.; Chorley, B.N.; Carswell, G.; Jones, W.; Wehmas, L.C.; Wood, C.E. Editor’s Highlight: Dose-Response Analysis of RNA-Seq Profiles in Archival Formalin-Fixed Paraffin-Embedded Samples. Toxicol. Sci. 2016, 154, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Wulfkuhle, J.D.; Calvert, V.S.; VanMeter, A.; Zhou, W.; Coukos, G.; Geho, D.H.; Petricoin, E.F.; Liotta, L.A. Laser-capture microdissection. Nat. Protoc. 2006, 1, 586–603. [Google Scholar] [CrossRef] [PubMed]
- Clemente, M.; Pérez-Alenza, M.D.; Illera, J.C.; Peña, L. Histological, immunohistological, and ultrastructural description of vasculogenic mimicry in canine mammary cancer. Vet. Pathol. 2010, 47, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Vincek, V.; Nassiri, M.; Knowles, J.; Nadji, M.; Morales, A.R. Preservation of Tissue RNA in Normal Saline. Lab. Investig. 2003, 83, 137–138. [Google Scholar] [CrossRef] [PubMed]
- Kowalewski, M.P.; Kautz, E.; Högger, E.; Nadji, M.; Morales, A.R. Interplacental uterine expression of genes involved in prostaglandin synthesis during canine pregnancy and at induced prepartum luteolysis/abortion. Reprod. Biol. Endocrinol. 2014, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Iussich, S.; Maniscalco, L.; Di Sciuva, A.; Iotti, B.; Morello, E.; Martano, M.; Gattino, F.; Buracco, P.; De Maria, R. PDGFRs expression in dogs affected by malignant oral melanomas: Correlation with prognosis. Vet. Comp. Oncol. 2016. [CrossRef] [PubMed]
- Kodama, A.; Sakai, H.; Matsuura, S.; Murakami, M.; Murai, A.; Mori, T.; Maruo, K.; Kimura, T.; Masegi, T.; Yanai, T. Establishment of canine hemangiosarcoma xenograft models expressing endothelial growth factors, their receptors, and angiogenesis-associated homeobox genes. BMC Cancer 2009, 9, 363. [Google Scholar] [CrossRef] [PubMed]

| Case # | Gender | Breed | Age (Years) | Subtype of Simple Carcinoma | Age of Sample (Months) |
|---|---|---|---|---|---|
| 1 | f | Basset | 12 | tubular | 3 |
| 2 | f | Vizsla | 10 | cystic-papillary | 18 |
| 3 | f | Samoyed | 5 | tubulo-papillary | 7 |
| 4 | f | Maltese | 14 | tubular | 3 |
| 5 | f | Tibetan Terrier | 12 | tubular | 15 |
| 6 | f/n | West Highland White Terrier | 12 | tubular-solid | 13 |
| 7 | f | Havanese | 13 | tubular | 11 |
| 8 | f | Chihuahua | 8 | tubulo-papillary | 7 |
| 9 | f/n | Bracke | 9 | cribriform | 14 |
| 10 | f/n | n.d. | 13 | tubular | 7 |
| 11 | f/n | Appenzell Mountain Dog | 6 | tubular | 18 |
| 12 | f | Boxer | 9 | tubulo-papillary | 8 |
| 13 | f | n.d. | 4 | cystic-papillary | 23 |
| Gene Name | Protein Name | Function | Expression in CAS | qPCR | IHC | Ref. |
|---|---|---|---|---|---|---|
| PDGFRB | PDGFRβ (Platelet-derived growth factor beta) | Cell-surface tyrosine kinase receptor | upregulated | × | × | [17,18,19] |
| MMP2 | MMP2 (Matrix metalloproteinase 2) | Metalloproteinase | upregulated | × | × | [20,21,22,23] |
| COL1A1 | Col1a1 (Collagen 1 α 1) | Extracellular matrix | upregulated | × | [22,23,24,25,26] | |
| FAP | FAP, Fibroblast activation protein | Serine protease | upregulated | × | [27,28] | |
| ACTA2 | αSMA (α smooth muscle actin, aorta) | Cytoskeleton | upregulated | × | × | [29,30,31,32,33,34] |
| CXCL12 | SDF1 (Stromal cell-derived factor 1) | Chemokine | upregulated | × | × | [20,35,36] |
| IL6 | IL-6 (Interleukin-6) | Cytokine | upregulated | × | [37,38] | |
| FGF2 | bFGF (Basic fibroblast growth factor) | Growth factor | upregulated | × | [2,39] | |
| CAV1 | Cav1 (Caveolin-1) | Possibly stabilisation of caveolar membranes | Downregulated * | × | [2,40] |
| Cresyl Violet Staining for FFPE Tissue Sections | |
|---|---|
| 100% Xylene, bath 1 | 5 min |
| 100% Xylene, bath 2 | 5 min |
| 100% Ethanol | 30 s |
| 95% Ethanol | 30 s |
| 70% Ethanol | 30 s |
| dH2O | 10 s |
| Cresyl violet (75% Ethanol with Diethylpyrocarbonate (DEPC) treated dH2O, pH 8.0) | 15 s |
| dH2O | 10 s |
| 70% Ethanol | 10 s |
| 95% Ethanol, bath 1 | 10 s |
| 95% Ethanol, bath 2 | 10 s |
| 100% Ethanol, bath 1 | 30–60 s |
| 100% Ethanol, bath 2 | 30–60 s |
| Gene Target | Sequence | Amplicon Length (nt) | Taqman® Order Number or Reference |
|---|---|---|---|
| cGAPDH | Fw: 5′-GCTGCCAAATATGACGACATCA-3′ Rev: 5′-GTAGCCCAGGATGCCTTTGAG-3′ Probe: 5′-TCCCTCCGATGCCTGCTTCACTACCTT-3′ | 75 | [63] |
| cPPIA | Manufacturer’s proprietary information | 92 | Cf03986523_gH |
| cB2M | Manufacturer’s proprietary information | 87 | Cf02659077_m1 |
| cPDGFRB | Manufacturer’s proprietary information | 60 | Cf02626568_g1 |
| cMMP2 | Manufacturer’s proprietary information | 58 | Cf02623423_m1 |
| cCOL1A1 | Manufacturer’s proprietary information | 87 | Cf02623126_m1 |
| cFAP | Manufacturer’s proprietary information | 69 | Cf02657429_m1 |
| cACTA2 | Manufacturer’s proprietary information | 86 | Cf02668774_mH |
| cCXCL12 | Manufacturer’s proprietary information | 86 | Cf02625258_m1 |
| cIL6 | Manufacturer’s proprietary information | 68 | Cf02624151_m1 |
| Antibody | Source, Order Information | Type | Dilution | Reference |
|---|---|---|---|---|
| PDGFR-β | Santa Cruz Biotechnology, sc-432 | Rabbit polyclonal | 1:50 | [64] |
| FAP | Abcam, ab53066 | Rabbit polyclonal | 1:100 | [50] |
| MMP-2 | Thermo Scientific, Ab-7, #RB-1537-P1 | Rabbit polyclonal | 1:100 | [53] |
| αSMA | Dako, Clone 1A4, Code M0851 | Mouse monoclonal | 1:400 | - |
| SDF1 | Abcam, ab9797 | Rabbit polyclonal | 1:100 | - |
| FGF-2 | Santa Cruz Biotechnology, (147) sc-79 | Rabbit polyclonal | 1:100 | [65] |
| Caveolin-1 | Santa Cruz Biotechnology, (N-20) sc-894 | Rabbit polyclonal | 1:100 | [55] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ettlin, J.; Clementi, E.; Amini, P.; Malbon, A.; Markkanen, E. Analysis of Gene Expression Signatures in Cancer-Associated Stroma from Canine Mammary Tumours Reveals Molecular Homology to Human Breast Carcinomas. Int. J. Mol. Sci. 2017, 18, 1101. https://doi.org/10.3390/ijms18051101
Ettlin J, Clementi E, Amini P, Malbon A, Markkanen E. Analysis of Gene Expression Signatures in Cancer-Associated Stroma from Canine Mammary Tumours Reveals Molecular Homology to Human Breast Carcinomas. International Journal of Molecular Sciences. 2017; 18(5):1101. https://doi.org/10.3390/ijms18051101
Chicago/Turabian StyleEttlin, Julia, Elena Clementi, Parisa Amini, Alexandra Malbon, and Enni Markkanen. 2017. "Analysis of Gene Expression Signatures in Cancer-Associated Stroma from Canine Mammary Tumours Reveals Molecular Homology to Human Breast Carcinomas" International Journal of Molecular Sciences 18, no. 5: 1101. https://doi.org/10.3390/ijms18051101
APA StyleEttlin, J., Clementi, E., Amini, P., Malbon, A., & Markkanen, E. (2017). Analysis of Gene Expression Signatures in Cancer-Associated Stroma from Canine Mammary Tumours Reveals Molecular Homology to Human Breast Carcinomas. International Journal of Molecular Sciences, 18(5), 1101. https://doi.org/10.3390/ijms18051101