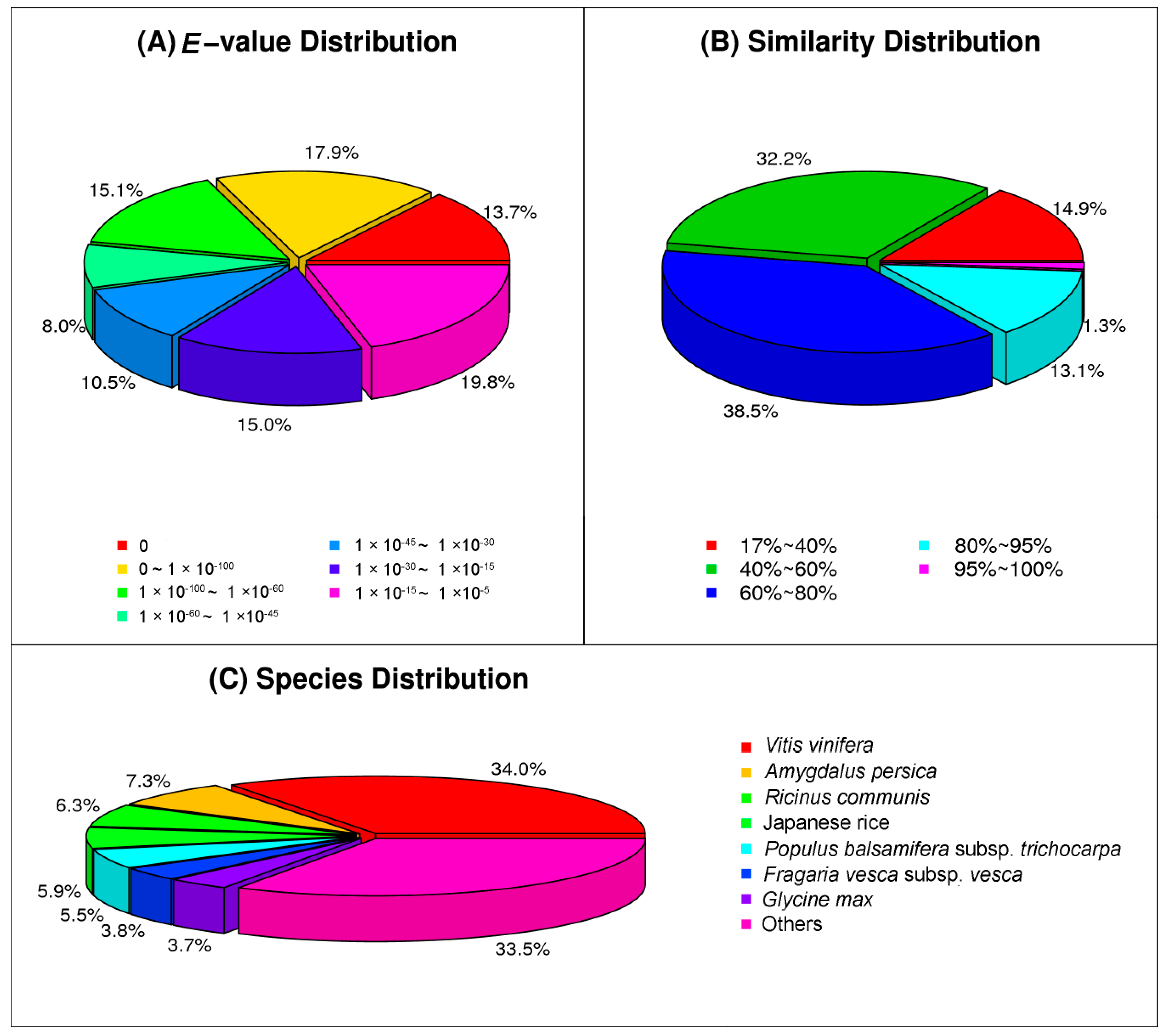
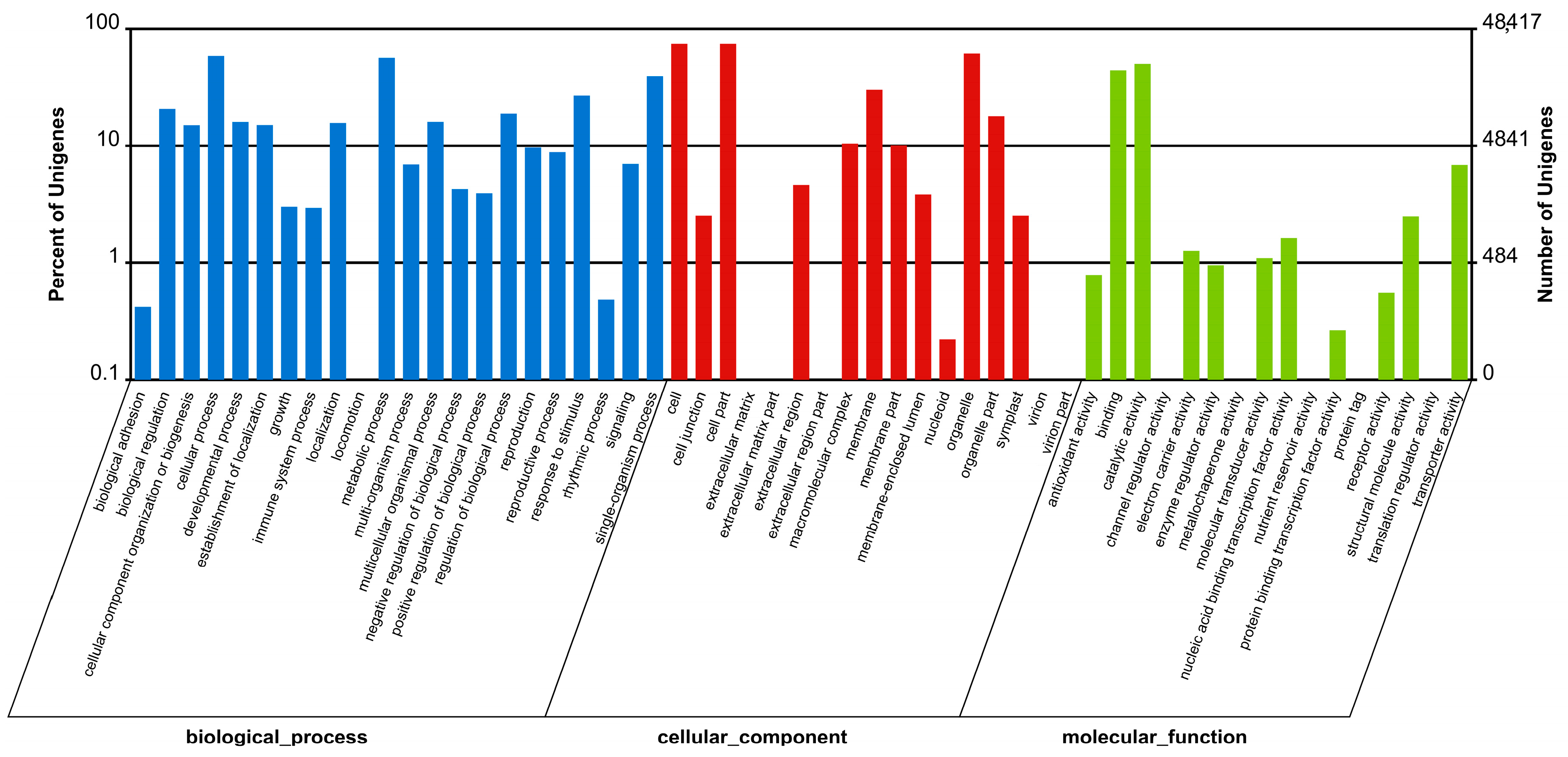
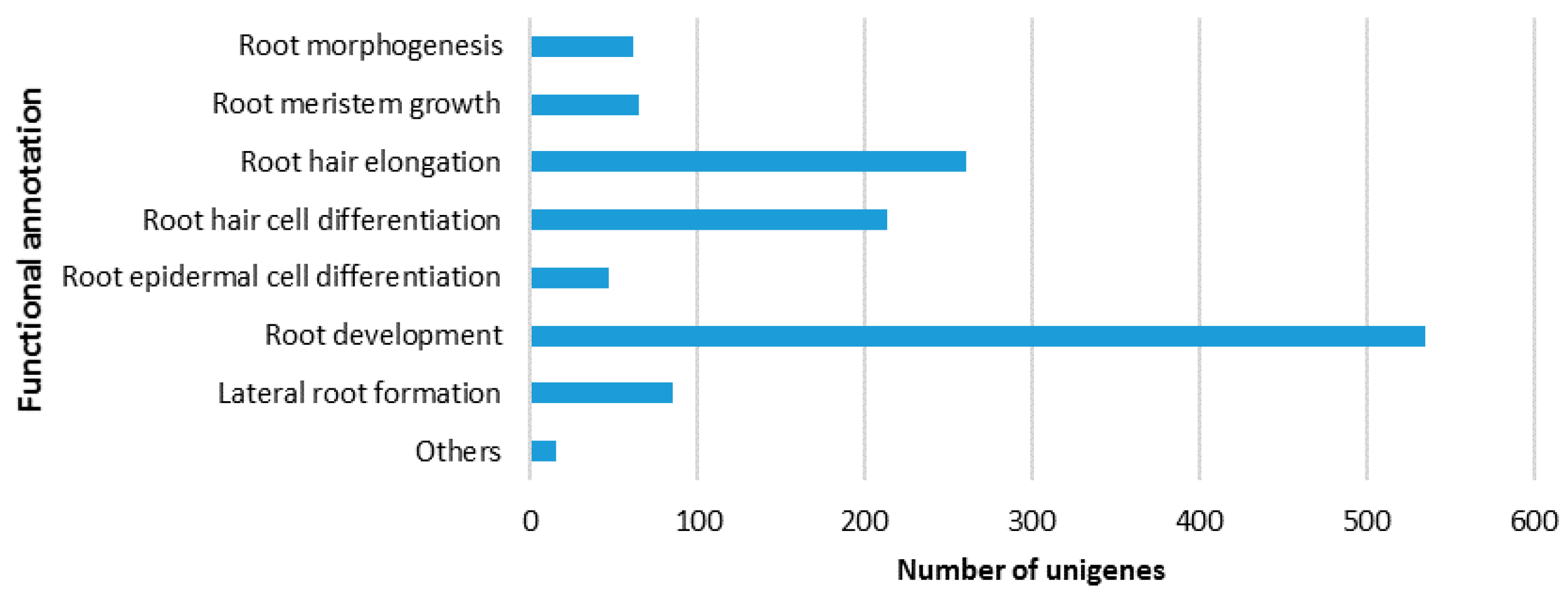
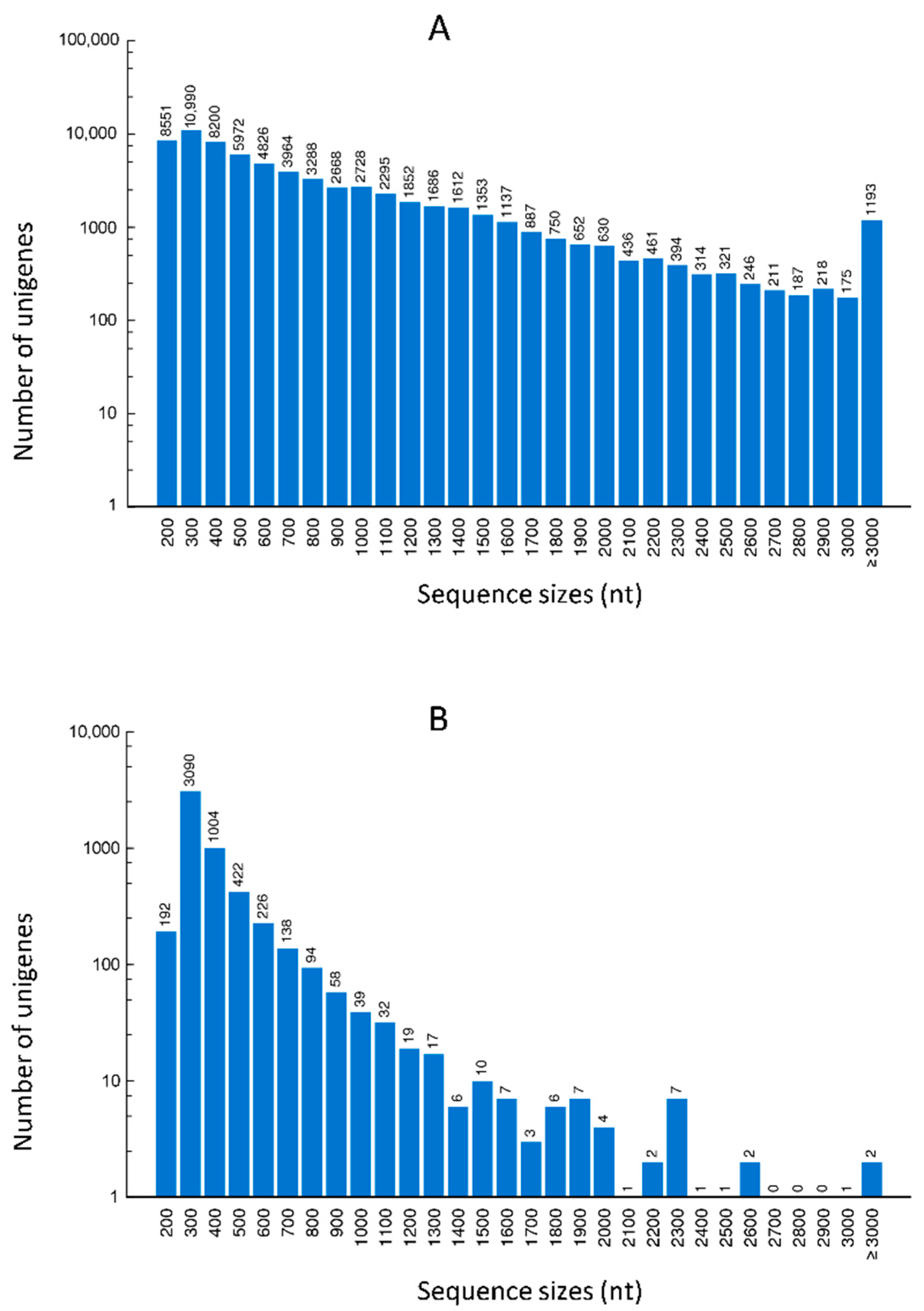
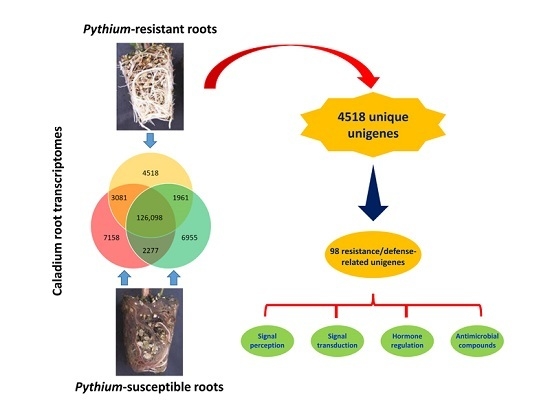
In response to attacks by necrotrophic pathogens, the innate immune system of plants perceives PAMPs with PRRs and activates PAMP-triggered immunity (PTI) against the pathogens [
13]. PTI involves a number of cellular processes, including accumulation of reactive oxygen species and nitric oxide, modulation of hormone level, secretion of antimicrobial compounds, and reinforcing of the cell wall [
34]. In this study, we compared unigenes in the root transcriptome of “Candidum” (moderately resistant to
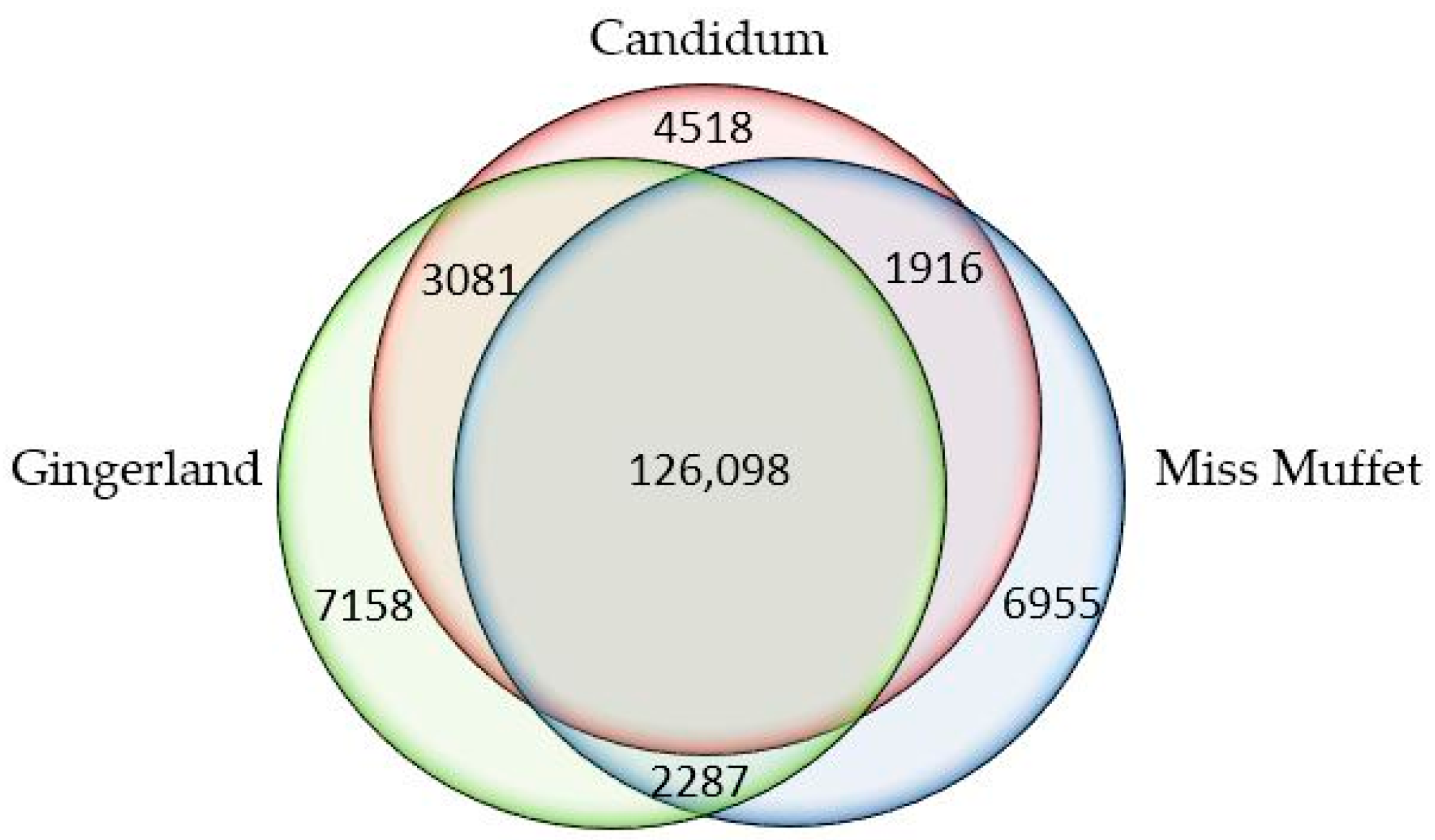
P. myriotylum) with those in the root transcriptomes of “Gingerland” and “Miss Muffet” (both highly susceptible to the pathogen) and identified 4518 unigenes that were present only in the root transcriptome of “Candidum” (
Figure 6 and
Table S7). These unique unigenes were examined for potential involvement in disease resistance and plant defense, in particular, (1) early signal perception and transduction in plant-pathogen interactions; (2) hormone regulation during plant defense responses to necrotrophic pathogens; and (3) secondary metabolites with antimicrobial activities in plant defense (
Table 5).
2.3.1. Early Signal Perception and Transduction in Plant-Pathogen Interactions
Recognition of PAMPs through membrane-localized PRRs during plant-pathogen interactions can induce an immune response syndrome [
35]. Previous studies have indicated that a series of plant RLKs could be PRRs for plant immune responses [
36]. Lack of RLKs could attenuate plant responses to pathogens, resulting in disease susceptibility [
37]. All RLKs are transmembrane proteins, each containing a signal sequence, a putative extracellular domain at the amino-terminal, and an intracellular kinase domain at carboxyl-terminal [
38]. Previous studies have shown that RLKs participate in many biologically important processes that confer disease resistance [
39].
Of the 4518 unigenes found only in “Candidum”, 50 unigenes can be translated into RLKs, including one
l-type lection receptor kinase (LecRK), one cysteine-rich receptor-like kinase (CRK2), 12 receptor-like serine/threonine-protein kinases (STKs), 31 proline-rich receptor-like protein kinases (PERKs), and five possible RLKs (
Table 5 and
Table S8). LecRKs are important immune receptors that bind to the effectors produced by
Phytophthora infestans [
40]. Previous studies have shown that
LecRK genes confer resistance in
Arabidopsis to several necrotrophic oomycetes including
Phytophthora brassicae and
Botrytis cinerea [
41,
42]. The one
LecRK homolog in “Candidum” may play a positive role in its disease resistance. CRKs are one of the largest classes of RLKs in plants [
43]. Studies in
Arabidopsis have shown that CRKs regulate plant development, stress adaptation, and disease resistance [
44]. Yang et al. [
45] found that a novel
CRK homolog (
TaCRK1) is involved in wheat resistance to
Rhizocotonia cerealis and the expression levels of
TaCRK1 were significantly higher in moderately resistant lines than in highly susceptible lines. In “Candidum” root transcriptome, only one
CRK homolog (
CRK2) was observed (
Table 5). Another important type of plant receptor-like kinases are STKs, which can phosphorylate the hydroxyl group (OH) of serine or threonine residues, resulting in functional alterations of target proteins [
46]. The phosphorylated proteins play an important role in disease resistance pathway by triggering a series of downstream activities including protein modulation, protein-protein interaction, transcription factor activation, and secretion of antimicrobial metabolites [
47]. It has been shown that two wheat
STK homologs,
Tsn1 and
Stpk-V, are the key genes conferring resistance against biotrophic
Blumeria graminis (wheat powdery mildew) and the necrotrophic pathogens
Stagonospora nodorum (Stagonospora nodorum blotch) and
Pyrenophora tritici-repentis (tan spot) [
46,
48]. In this study, 12
STK homologs were observed exclusively in the “Candidum” root transcriptome (
Table 5). In addition, we found that a majority of RLKs in the “Candidum” (60%) root transcriptome belong to PERKs. In
Arabidopsis, PERKs represent a small family of RLKs, all sharing an extracellular domain, a transmembrane domain, and a kinase domain [
38]. PERKs in
Arabidopsis roots are involved in callose and cellulose deposition; PERK1 mutants have lost their ability to accumulate callose and cellulose in cell walls [
49]. Silva and Goring [
50] found that PERK1 could response to both mechanical stresses and infection by fungal pathogens and may have a function in the general perception of wounds or recognition of pathogen stimuli. In the “Candidum” root transcriptome, 31
PERK homologs were observed (
Table 5), and these
PERKs may be good candidates for further gene expression studies. There were five possible
RLK-like unigenes (
Table 5) that could not be further annotated, suggesting that these
RLK-like unigenes may represent unique transcripts in caladium and their functions remain to be studied.
After plant perception of PAMPs, signals from the PRR complexes are transferred to downstream targets through several secondary messengers, including influx of calcium (Ca
2+), reactive oxygen species (ROS) burst, and activation of mitogen-activated protein kinases (MAPKs) [
51]. The calcium influx is an early step in the signal transduction processes [
52]. Ca
2+ influx can be sensed by several stimulus-specific Ca
2+-binding sensors such as calcium-dependent protein kinases (CPKs) and Calmodulins (CaMs). Previous studies have shown that transgenic potato plants containing
StCDPK4 and
StCDPK5 (
CPK homologs) gain resistance to
Phytophthora infestans [
53]. A wheat
CPK homolog (
TaCPK7-D) is involved in determining resistance to necrotrophic pathogen
Rhizoctonia cerealis. Here, we identified eight CPKs homologs only in “Candidum“ (
Table 5 and
Table S9) and suspect that they may be important signal sensors which modulate resistance to necrotrophic pathogens. CaMs and CaM-like (CML) genes could trigger plant defense responses to a broad range of pathogens. In tobacco, a
CML gene,
NtCaM13, is a key gene for resistance to the necrotrophic pathogens
Pythium aphanidermatum and
Rhizoctonia solani [
54]. Four
CaM gene homologs were identified among “Candidum” unigenes (
Table 5). ROS are important signals to modulate the expression of defense-related genes to fungal pathogen attacks [
55]. The production of ROS involves a number of oxidative enzymes such as the NADPH (nicotinamide adenine dinucleotide phosphate) oxidase, the superoxide dismutase, the cell wall peroxidase, the oxalate oxidase, the lipoxygenase, and the polyamine oxidase [
56]. These enzymes work collaboratively or separately to initiate a series of responses including cell wall remodeling, signal transduction, programming cell death, and post-translation regulation to resist pathogens [
56]. So far, our understanding of the roles ROS play in plant roots and resistance against necrotrophic pathogens is still rudimentary [
56]. In banana, studies have shown that a NADPH oxidase homolog (
Rboh) confers resistance against the necrotrophic pathogen
Fusarium oxysporum. In
Arabidopsis and cotton,
F. oxysporum-resistant lines accumulated ROS faster than the susceptible lines [
57,
58]. In this study, we identified five putative ROS-related unigenes (superoxide dismutase) that were exclusively expressed in “Candidum” (
Table 5). MAPK activation is another early signaling activity after plants sense the attack from pathogens [
59]. Plant MAPK cascades transfer signals from PRRs to downstream components [
60]. In
Arabidopsis, MAPK cascades involve several MAPKs such as MEKK1, MKK1/MKK2 (two redundant MAPKKs), MPK4/MPK5 (two redundant MAPKs), and MPK3/MPK6 (two partially redundant MAPKs). Previous studies have shown that MPK3, MPK4, MPK6, MKK1, MKK2, and MEKK1 are involved in innate immune responses against necrotrophic pathogens [
61]. Of 4956 unigenes only observed in the “Candidum” root transcriptome, there were eight
MAPK homologs including three
MPK1, two
MPK2, one
MPK4, one
MPK5, and one
MPK14 homolog (
Table 5).
2.3.2. Hormone Regulation during Plant-Pathogen Interactions
Increased biosynthesis of hormones is an important plant defense response to invading pathogens [
37]. In plants, ethylene (ET) and jasmonic acid (JA) are two important hormones involved in resistance against necrotrophic pathogens.
S-adenosylmethionine (
S-AdoMet) and 1-aminocyclopropane-1-carboxylic acid (ACC) are two precursors of ethylene [
62]. Enzymes catalyzing ET biosynthesis reactions include ACC synthase (ACS) and ACC oxidase (ACO) [
62]. Among the annotated unigenes in “Candidum”, both key enzymes participating in ET synthesis were identified (
Table 5 and
Table S10). Synthesized ethylene can be then perceived and transduced by ethylene receptors to trigger downstream biological responses. There are several types of ethylene receptors in
Arabidopsis, such as ethylene receptors (ETRs), ethylene response sensors (ERSs), and ethylene insensitive (EINs) [
63]. However, the roles ethylene receptors play in necrotrophic pathogen resistance seem to be different depending on plant species and the types of pathogens. In
Arabidopsis, the
ein2 (ethylene insensitive) mutant lines displayed increased susceptibility to the necrotrophic pathogen
Botrytis cinerea [
64], while in soybean, the
etr1 and
etr2 (ethylene-resistant) mutant lines showed increased resistance against
Phytophthora sojae [
65]. In this study, we observed 14
ETR homologs that were expressed in the three caladium cultivars. One
ETR-like unigene was expressed exclusively in the “Candidum” root transcriptome; this unigene could be a good candidate for future studies.
JA is another important hormone in plant defense against many necrotrophic pathogens [
66]. It has been reported that exogenous applications of methyl jasmonate to
Arabidopsis enhance plant resistance to root rot disease caused by the necrotrophic pathogen
Pythium mastophorum [
67]. Cohen et al. [
68] found that methyl jasmonate-treated potatoes showed increased resistance to
Phytophthora infestans. Studies in tomato have shown that the
CORONATINE-INSENSITIVE1 gene (
COI1), the acyl-CoA oxidase gene (
ACX1A), the
suppressor of prosystemin-mediated responses2 (
SPR2), and the
defenseless1 gene (
def1) are involved in JA-induced defense responses against necrotrophic pathogens such as
Botrytis cinerea [
37]. Tomato plants with either one of these genes mutated could not correctly process JA synthesis, and JA signal perception or transduction, resulting in disrupted immune responses and susceptible plants [
37]. We found that 55 unigenes were potentially involved in JA synthesis and expressed in all three caladium cultivars. One
ACX1A homolog was expressed only in “Candidum” (
Table 5), which suggests that this unigene may be a good candidate for further gene expression studies.
2.3.3. Secondary Metabolites with Antimicrobial Activities
Plants can secrete secondary metabolites around their rhizosphere to inhibit or repel soil-borne pathogens. In general, phenolics and terpenoids are two primary classes of root-secreted antimicrobial metabolites [
69]. Common root-secreted phenolic compounds include phenylpropanoids and flavonoids [
70]. Many phenylpropanoids from a broad range of plant species have shown antimicrobial activities [
71]. Phenylpropanoids in chickpea roots affect resistance against the necrotrophic pathogen
Fusarium oxysporum; resistant chickpea varieties produce significantly more phenylpropanoids than susceptible ones [
72]. Lanoue et al. [
71] found that phenylpropanoids were constitutively secreted by the barley root system and inhibited
Fusarium graminearum spore germination. In “Candidum“, we found two unigenes that are likely involved in phenylpropanoid synthesis. Flavonoids are often one of the largest groups of phenolics in root exudates [
69]. Studies in maize have indicated that high levels of flavonoids are associated with an increased defensive state in its roots [
73]. We identified four unigenes in “Candidum” that are likely involved in flavonoid synthesis (
Table 5 and
Table S11).
Terpenoids are another large group of compounds in root exudates that have antimicrobial activities [
74]. Some volatile terpenoids emitted from roots could act as a direct barrier against pathogens [
69]. In
Arabidopsis, Steeghs et al. [
75] found a below-ground volatile compound (monoterpene 1,8-cineole) which had direct effects on
Pseudomonas syringae. Kapulnik et al. [
76] observed that strigolactones released from roots inhibited the growth of several soil-borne pathogens, including
Fusarium oxysporum,
Fusarium solani, and
Macrophomina phaseolina. We identified four unigenes in the “Candidum” root transcriptome that are likely involved in the terpenoid synthesis pathway. Derivatives of tryptophan are another important antimicrobial root exudates. Typical examples of tryptophan derivatives include camalexin, which is produced by
Arabidopsis and exhibits antimicrobial activities to a wide range of soil-borne pathogens including
Pythium sylvaticum,
Rhizoctonia solani, and
Sclerotinia sclerotiorum [
77,
78]. In the “Candidum” root transcriptome, we found nine tryptophan synthesis-related unigenes (
Table 5 and
Table S11), which may be involved in resistance against soil-borne necrotrophic pathogens. One of these unigenes is root-specific.