Identification of a Ribose-Phosphate Pyrophosphokinase that Can Interact In Vivo with the Anaphase Promoting Complex/Cyclosome
Abstract
:1. Introduction
2. Results
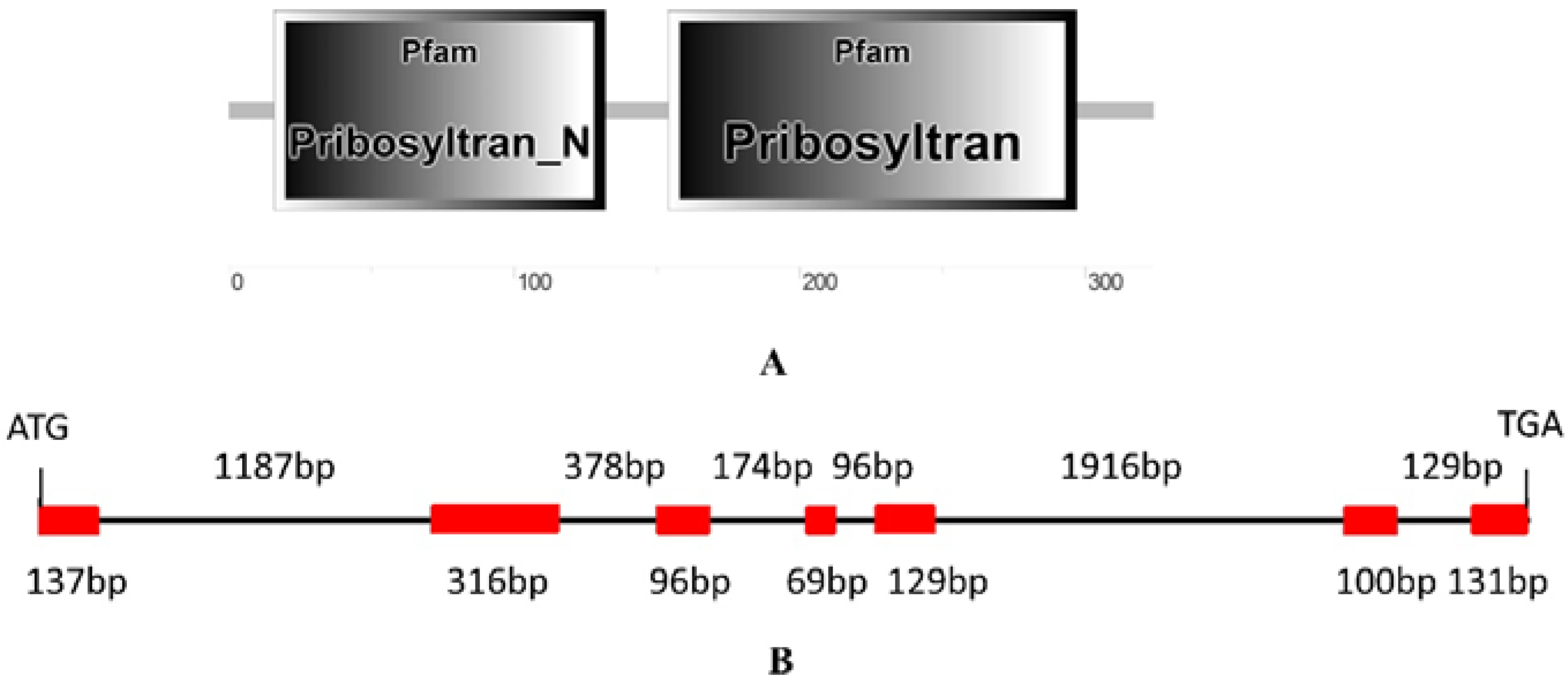
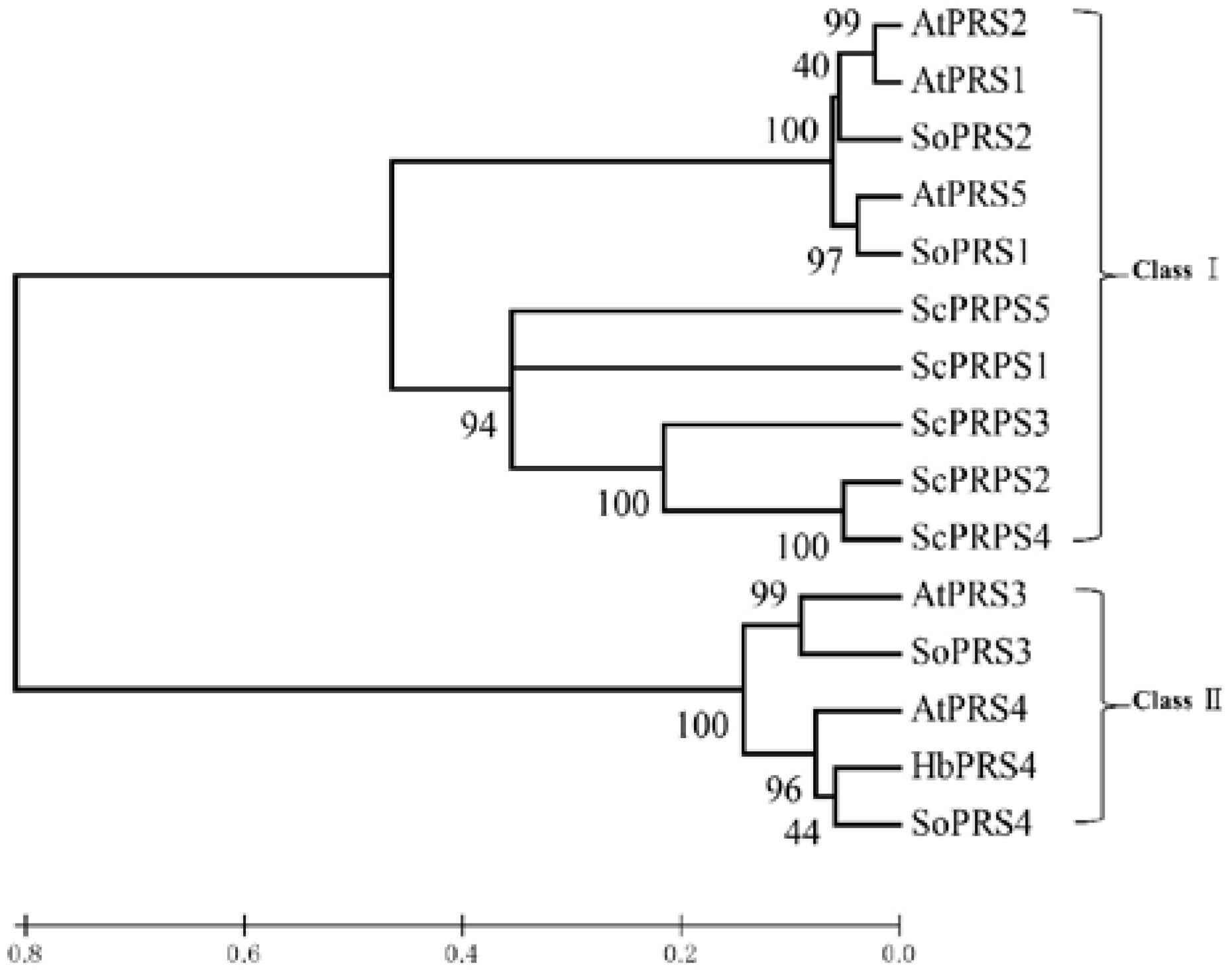
2.1. Identification and Isolation of Rubber Tree PRS4
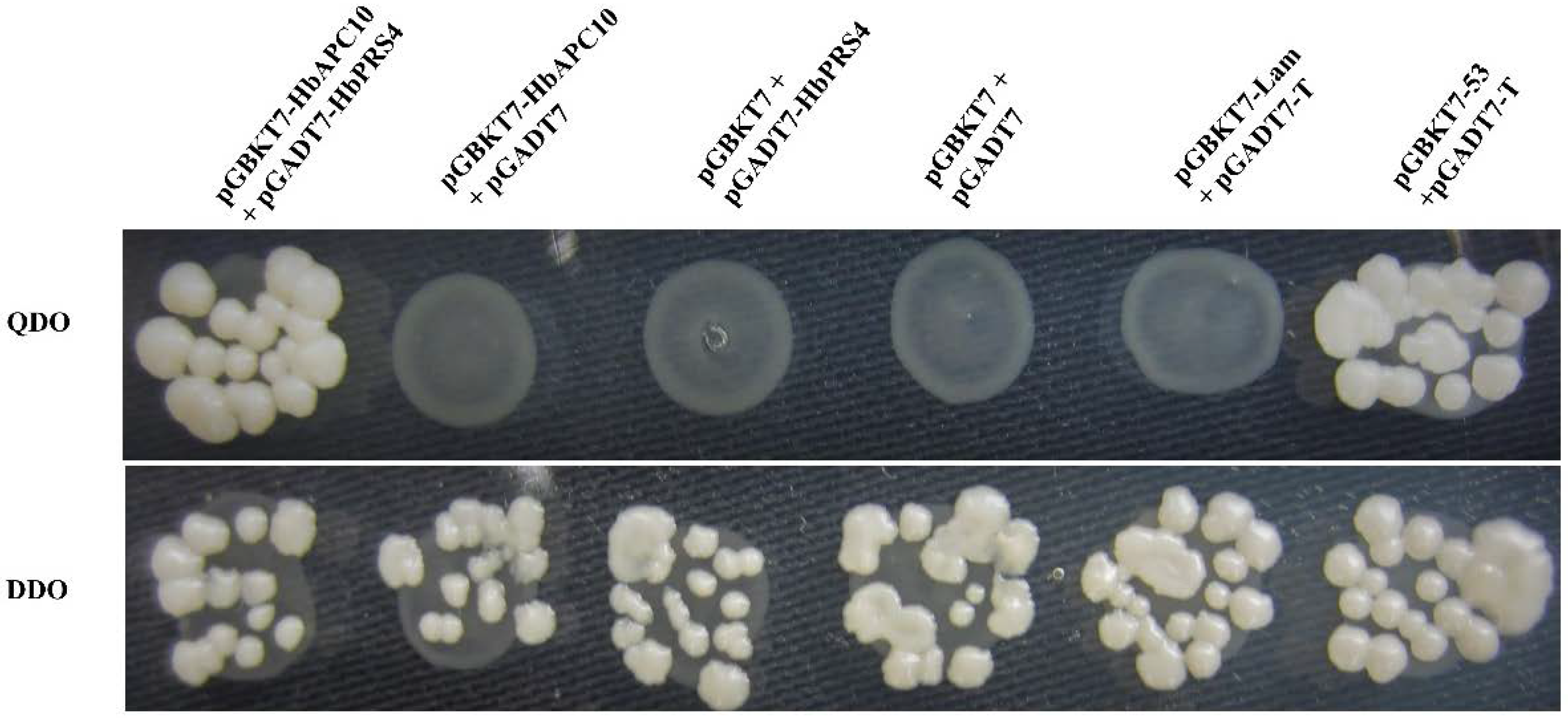
2.2. Confirmation of the Interaction of HbAPC10 with HbPRS4 by Yeast Two-Hybrid Assays
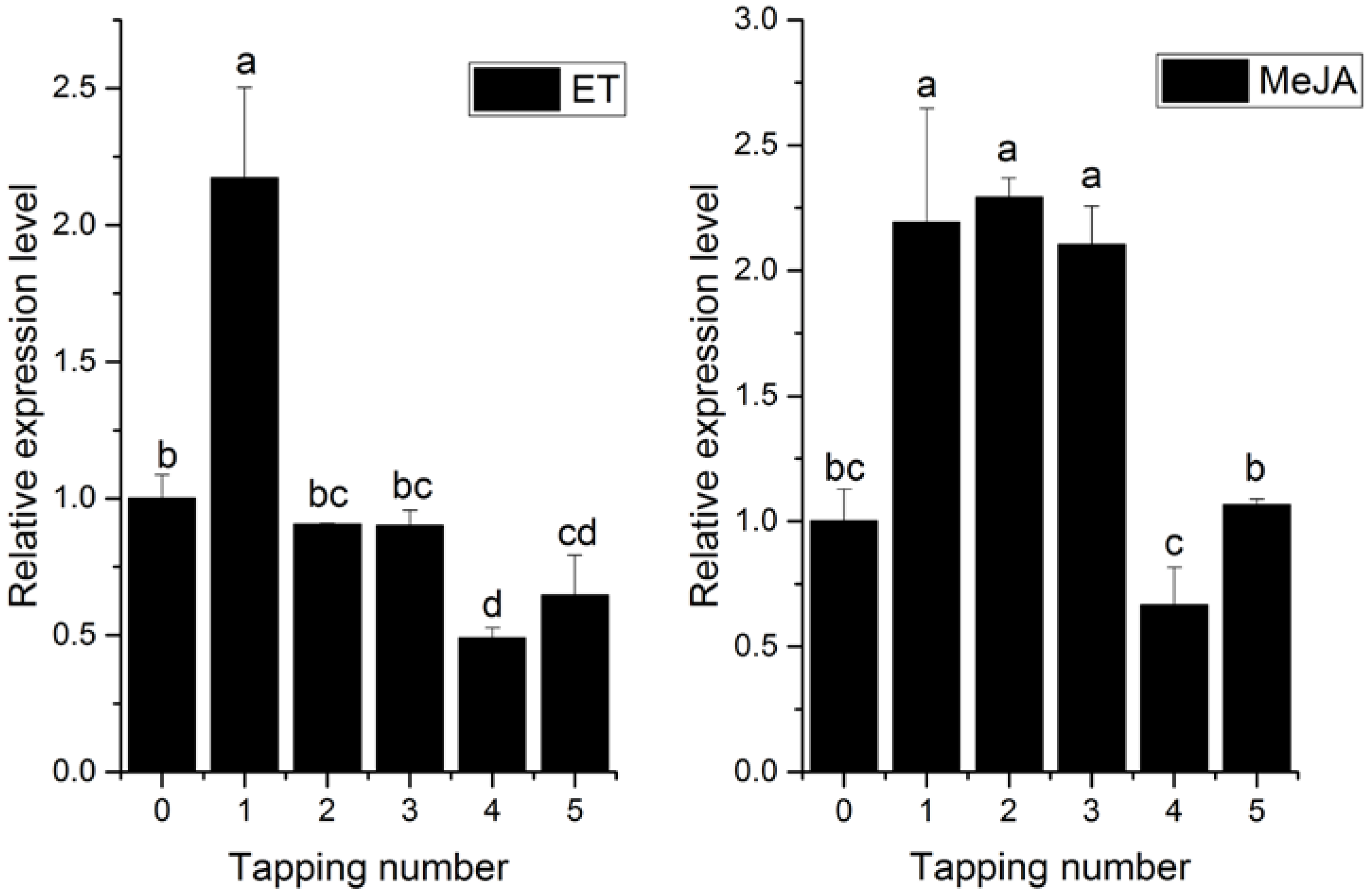
2.3. Transcriptional Profiles of HbPRS4 in Different Tissues and in Response to Different Stimulus
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Yeast Two-Hybrid Screening
4.3. Isolation of HbPRS4 and Bioinformatic Analysis
4.4. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PRS4 | Ribose-phosphate pyrophosphokinase 4 |
| PRPP | 5-Phospho-d-ribosyl-1-diphosphate |
| APC/C | Anaphase promoting complex/cyclosome |
| ET | Ethephon |
| MeJA | Methyl jasmonate |
| ABA | Abscisic acid |
| qRT-PCR | Real-time quantitative PCR |
| ORF | Open reading frame |
| TSA | Transcriptome shotgun assembly |
| QDO | SD/-Ade/-His/-Leu/-Trp |
| DDO | SD/-Leu/-Trp |
| PCR | Polymerase chain reaction |
| RT-PCR | Reverse transcription PCR |
| qRT-PCR | Quantitative real time PCR |
| ATP | Adenosine triphosphate |
| ADP | Adenosine diphosphate |
| AMP | Adenosine monophosphate |
| CT | Cycle threshold |
References
- Hove-Jensen, B. Mutation in the phosphoribosylpyrophosphate synthetase gene (PRS) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J. Bacteriol. 1988, 170, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Khorana, H.G.; Fernandes, J.F.; Kornberg, A. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J. Biol. Chem. 1958, 230, 941–948. [Google Scholar] [PubMed]
- Krath, B.N.; Hove-Jensen, B. Implications of secondary structure prediction and amino acid sequence comparison of class I and class II phosphoribosyl diphosphate synthases on catalysis, regulation, and quaternary structure. Protein Sci. 2001, 10, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Krath, B.N.; Hove-Jensen, B. Class II recombinant phosphoribosyl diphosphate synthase from spinach. Phosphate independence and diphosphoryl donor specificity. J. Biol. Chem. 2001, 276, 17851–17856. [Google Scholar] [CrossRef] [PubMed]
- Krath, B.N.; Hove-Jensen, B. Organellar and cytosolic localization of four phosphoribosyl diphosphate synthase isozymes in spinach. Plant Physiol. 1999, 119, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Kadziola, A.; Jepsen, C.H.; Johansson, E.; McGuire, J.; Larsen, S.; Hove-Jensen, B. Novel Class III phosphoribosyl diphosphate synthase: Structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J. Mol. Biol. 2005, 354, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Krath, B.N.; Eriksen, T.A.; Poulsen, T.S.; Hove-Jensen, B. Cloning and sequencing of cdnas specifying a novel class of phosphoribosyl diphosphate synthase in Arabidopsis thaliana. Biochim. Biophys. Acta 1999, 1430, 403–408. [Google Scholar] [CrossRef]
- Hernando, Y.; Parr, A.; Schweizer, M. PRS5, the fifth member of the phosphoribosyl pyrophosphate synthetase gene family in Saccharomyces cerevisiae, is essential for cell viability in the absence of either PRS1 or PRS3. J. Bacteriol. 1998, 180, 6404–6407. [Google Scholar] [PubMed]
- Taira, M.; Ishijima, S.; Kita, K.; Yamada, K.; Iizasa, T.; Tatibana, M. Nucleotide and deduced amino acid sequences of two distinct cDNAs for rat phosphoribosylpyrophosphate synthetase. J. Biol. Chem. 1987, 262, 14867–14870. [Google Scholar] [PubMed]
- Roessler, B.J.; Bell, G.; Heidler, S.; Seino, S.; Becker, M.; Palella, T.D. Cloning of two distinct copies of human phosphoribosylpyrophosphate synthetase cDNA. Nucleic Acids Res. 1990, 18, 193. [Google Scholar] [CrossRef] [PubMed]
- Taira, M.; Iizasa, T.; Shimada, H.; Kudoh, J.; Shimizu, N.; Tatibana, M. A human testis-specific mRNA for phosphoribosylpyrophosphate synthetase that initiates from a non-AUG codon. J. Biol. Chem. 1990, 265, 16491–16497. [Google Scholar] [PubMed]
- Hewitt, M.M.; Carr, J.M.; Williamson, C.L.; Slocum, R.D. Effects of phosphate limitation on expression of genes involved in pyrimidine synthesis and salvaging in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Pujarniscle, S. Caractère lysosomal des lutoides du latex d’Hevea brasiliensis. Physiol. Veg. 1968, 6, 27–46. [Google Scholar]
- D’Auzac, J.; Jacob, J.L.; Chrestin, H. Physiology of Rubber Tree Latex; CRC Press: Boca Raton, FL, USA, 1989. [Google Scholar]
- Abraham, P.D.; Wycherley, P.R.; Pakianathan, S.W. Stimulation of latex flow in Hevea brasiliensis by 4-amino-3,5,6-trichloropicolinic acid and 2-chloroethane-phosphonic acid. J. Rubber Res. Inst. Malays. 1968, 20, 291–305. [Google Scholar]
- Hao, B.; Wu, J. Laticifer differentiation in Hevea brasiliensis: Induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 2000, 85, 37–43. [Google Scholar] [CrossRef]
- Amalou, Z.; Bangratz, J.; Chrestin, H. Ethrel (ethylene releaser)-induced increased in the adenylate pool and transtonoplast ph within Hevea latex cells. Plant Physiol. 1992, 98, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Gallois, R.; Prevot, J.C.; Clement, A.; Jacob, J.L. Purification and characterization of phosphoribosylpyrophosphate synthetase from rubber tree latex. Plant Physiol. 1997, 115, 847–852. [Google Scholar] [CrossRef] [PubMed]
- McLean, J.R.; Chaix, D.; Ohi, M.D.; Gould, K.L. State of the APC/C: Organization, function, and structure. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 118–136. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Capron, A.; Okresz, L.; Genschik, P. First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 2003, 8, 83–89. [Google Scholar] [CrossRef]
- Passmore, L.A.; McCormack, E.A.; Au, S.W.; Paul, A.; Willison, K.R.; Harper, J.W.; Barford, D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003, 22, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, C.M.; Kirschner, M.W. The ken box: An APC recognition signal distinct from the D box targeted by CDH1. Genes Dev. 2000, 14, 655–665. [Google Scholar] [PubMed]
- Harkness, T.A.; Shea, K.A.; Legrand, C.; Brahmania, M.; Davies, G.F. A functional analysis reveals dependence on the anaphase-promoting complex for prolonged life span in yeast. Genetics 2004, 168, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.L.; Bonham-Smith, P.C.; Postnikoff, S.; Gray, G.R.; Harkness, T.A. A role for the anaphase promoting complex in hormone regulation. Planta 2011, 233, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Eloy, N.B.; de Freitas Lima, M.; van Damme, D.; Vanhaeren, H.; Gonzalez, N.; de Milde, L.; Hemerly, A.S.; Beemster, G.T.; Inze, D.; Ferreira, P.C. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 2011, 68, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Yu, Y.; Duan, J.; Liao, Z.; Xiong, G.; Meng, X.; Liu, G.; Qian, Q.; Li, J. Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat. Commun. 2012, 3, 750. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, D.; Dong, H.; Gu, S.; Cheng, Z.; Gong, J.; Qin, R.; Jiang, L.; Li, G.; Wang, J.L.; et al. Rice APC/C(TE) controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 2012, 3, 752. [Google Scholar] [CrossRef] [PubMed]
- Hernando, Y.; Carter, A.T.; Parr, A.; Hove-Jensen, B.; Schweizer, M. Genetic analysis and enzyme activity suggest the existence of more than one minimal functional unit capable of synthesizing phosphoribosyl pyrophosphate in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 12480–12487. [Google Scholar] [CrossRef] [PubMed]
- Ugbogu, E.A.; Wippler, S.; Euston, M.; Kouwenhoven, E.N.; de Brouwer, A.P.; Schweizer, L.M.; Schweizer, M. The contribution of the nonhomologous region of PRS1 to the maintenance of cell wall integrity and cell viability. FEMS Yeast Res. 2013, 13, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Ugbogu, E.A.; Wang, K.; Schweizer, L.M.; Schweizer, M. Metabolic gene products have evolved to interact with the cell wall integrity pathway in Saccharomyces cerevisiae. FEMS Yeast Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Koslowsky, S.; Riegler, H.; Bergmuller, E.; Zrenner, R. Higher biomass accumulation by increasing phosphoribosylpyrophosphate synthetase activity in Arabidopsis thaliana and Nicotiana tabacum. Plant Biotechnol. J. 2008, 6, 281–294. [Google Scholar] [CrossRef] [PubMed]
- De Faiÿ, E.; Ebant, C.; Jacob, J.L. Cytology and cytochemistry of the laticiferous system. In Physiology of the Rubber Tree Latex; D’Auzac, J., Jacob, J.L., Chrestin, H., Eds.; CRC Press: Boca Raton, FL, USA, 1989; pp. 15–29. [Google Scholar]
- D’Auzac, J.; Jacob, J.L. The composition of latex from Hevea brasiliensis as a laticiferous cytoplasm. In Physiology of the Rubber Tree Latex; D’Auzac, J., Jacob, J.L., Chrestin, H., Eds.; CRC Press: Boca Raton, FL, USA, 1989; pp. 59–96. [Google Scholar]
- Coupé, M.; Chrestin, H. Physico-chemical and biochemical mechanisms of hormonal (ethylene) stimulation. In Physiology of the Rubber Latex Tree; D’Auzac, J., Jacob, J.L., Chrestin, H., Eds.; CRC Press: Boca Raton, FL, USA, 1989; pp. 295–319. [Google Scholar]
- Qin, B.; Zheng, F.; Zhang, Y. Molecular cloning and characterization of a Mlo gene in rubber tree (Hevea brasiliensis). J. Plant Physiol. 2015, 175, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Zhang, Y.; Wang, M.; Qin, B. Construction of a normalized yeast two-hybrid cDNA library of the latex of rubber tree (Hevea brasiliensis müll. Arg.). Plant Physiol. J. 2016, 52, 312–316. (in Chinese). [Google Scholar]
- Li, B.; Fields, S. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 1993, 7, 957–963. [Google Scholar] [PubMed]
- Iwabuchi, K.; Li, B.; Bartel, P.; Fields, S. Use of the two-hybrid system to identify the domain of p53 involved in oligomerization. Oncogene 1993, 8, 1693–1696. [Google Scholar] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA 6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence (5′ to 3′) | Name | Sequence (5′ to 3′) |
|---|---|---|---|
| PRS4-F1 | GGGACTCTCCTTCCAGCTCT | PRS4-R1 | CGTGAAACACCAATGGACAA |
| APC10-EcoRI-F | CGGAATTCATGGCAACAGAGTCAT | APC10-BamHI-R | CGGGATCCTCATCTCACTGAAGAGT |
| PRS4-EcoRI-F2 | CGGAATTCATGGAAAAGAAGGCAGA | PRS4-XhoI-R2 | CCGCTCGAGTCATATTTGCAGAGCA |
| PRS4-QF3 | CCCTCTGCTAAAGCAACGTC | PRS4-QR3 | CCATGGGAAAATGATCCAAC |
| Actin-F | GATGTGGATATCAGGAAGG | Actin-R | CATACTGCTTGGAGCAAGA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Zhang, Y.; Zhang, D.; Lu, Y.; He, H.; Zheng, F.; Wang, M. Identification of a Ribose-Phosphate Pyrophosphokinase that Can Interact In Vivo with the Anaphase Promoting Complex/Cyclosome. Int. J. Mol. Sci. 2017, 18, 617. https://doi.org/10.3390/ijms18040617
Yu H, Zhang Y, Zhang D, Lu Y, He H, Zheng F, Wang M. Identification of a Ribose-Phosphate Pyrophosphokinase that Can Interact In Vivo with the Anaphase Promoting Complex/Cyclosome. International Journal of Molecular Sciences. 2017; 18(4):617. https://doi.org/10.3390/ijms18040617
Chicago/Turabian StyleYu, Haiyang, Yu Zhang, Dong Zhang, Yanxi Lu, Haixia He, Fucong Zheng, and Meng Wang. 2017. "Identification of a Ribose-Phosphate Pyrophosphokinase that Can Interact In Vivo with the Anaphase Promoting Complex/Cyclosome" International Journal of Molecular Sciences 18, no. 4: 617. https://doi.org/10.3390/ijms18040617





