Genetic Variants Contributing to Colistin Cytotoxicity: Identification of TGIF1 and HOXD10 Using a Population Genomics Approach
Abstract
:1. Introduction
2. Results
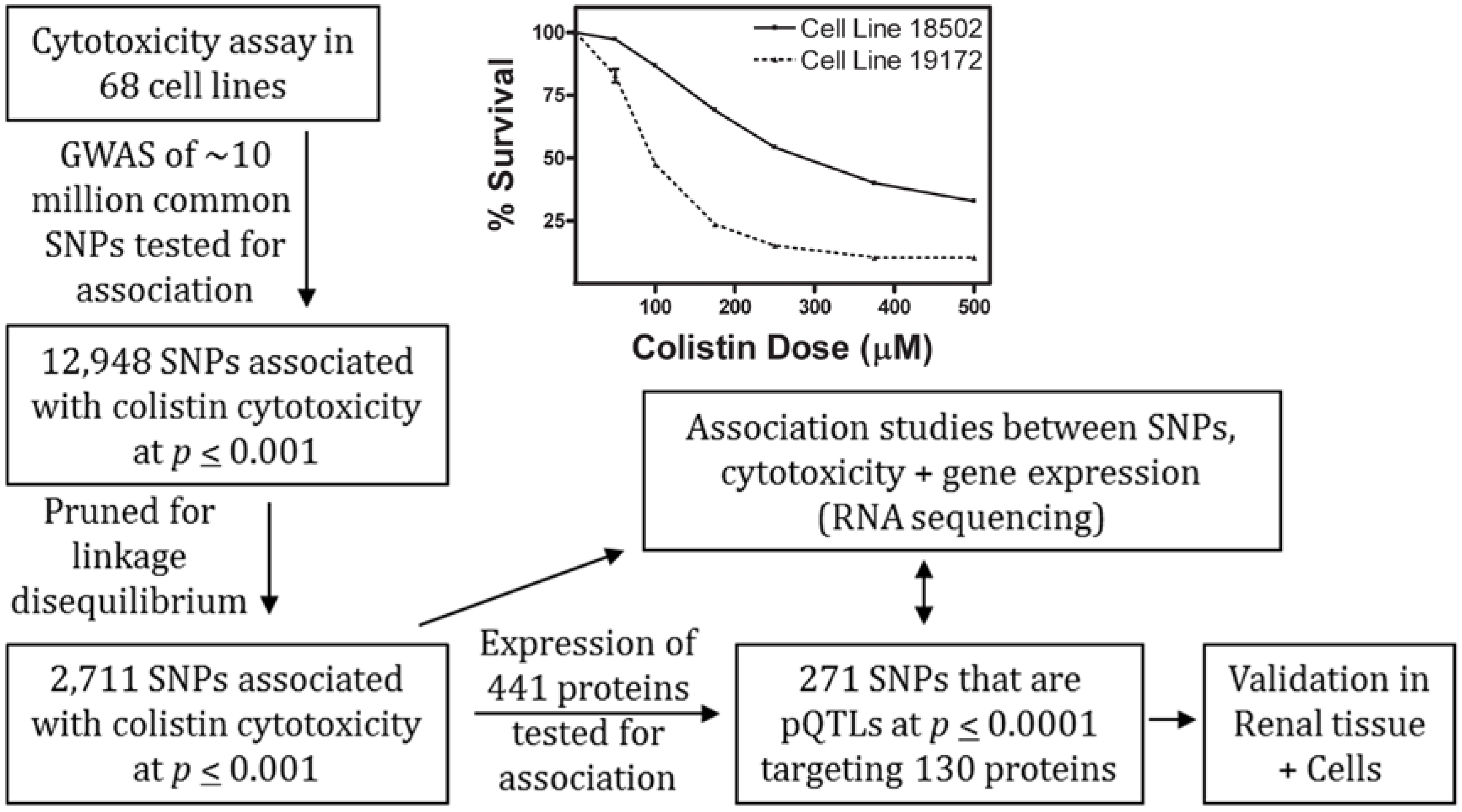
2.1. Colistin-Induced Cytotoxicity in Lymphoblastoid Cell Lines
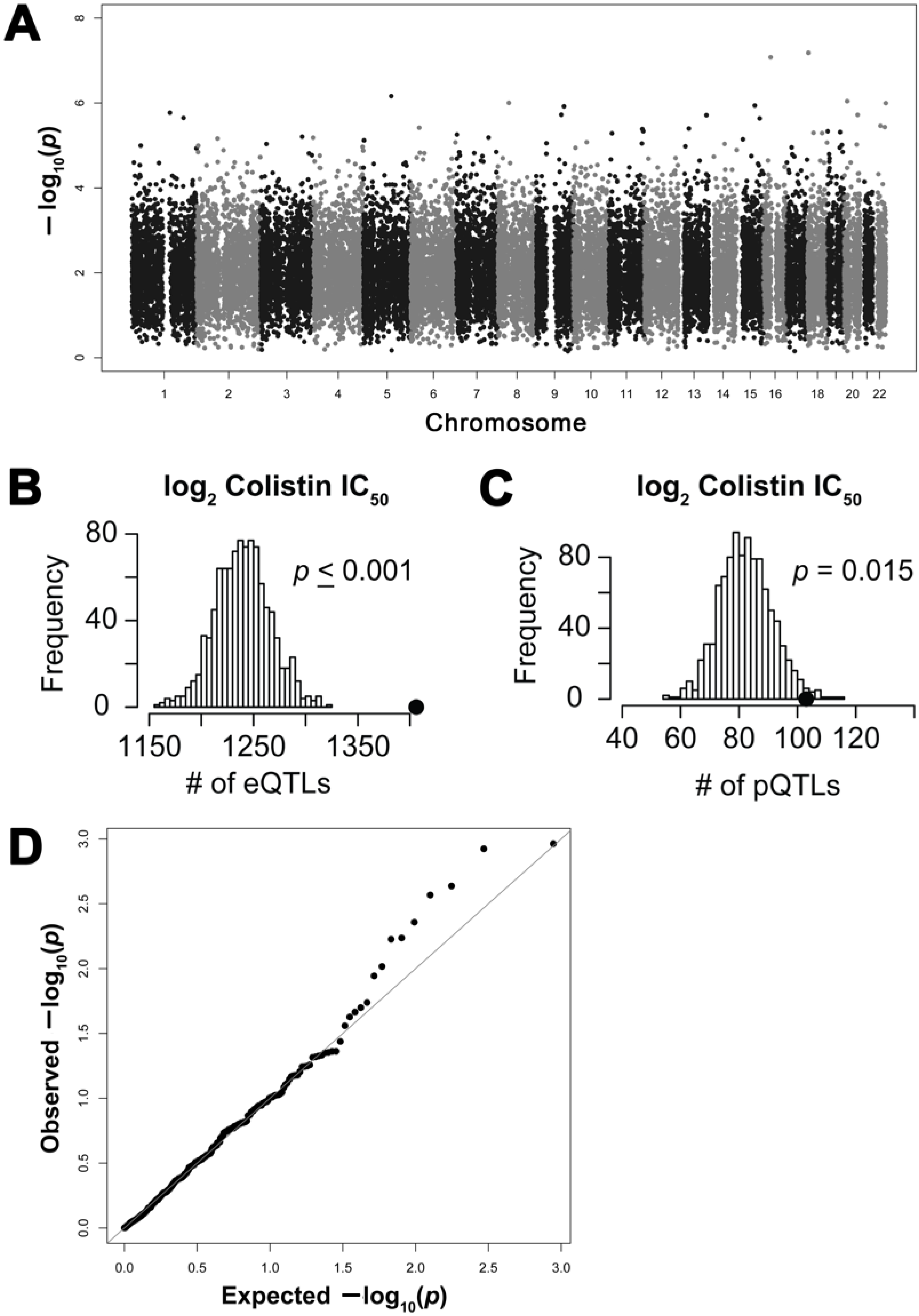
2.2. Genome-Wide Association Study of Colistin Cytotoxicity
2.3. Functional Enrichment of Expression Quantitative Trait Loci and Protein Quantitative Trait Loci
2.4. Association of Protein Expression with Colistin Cytotoxicity
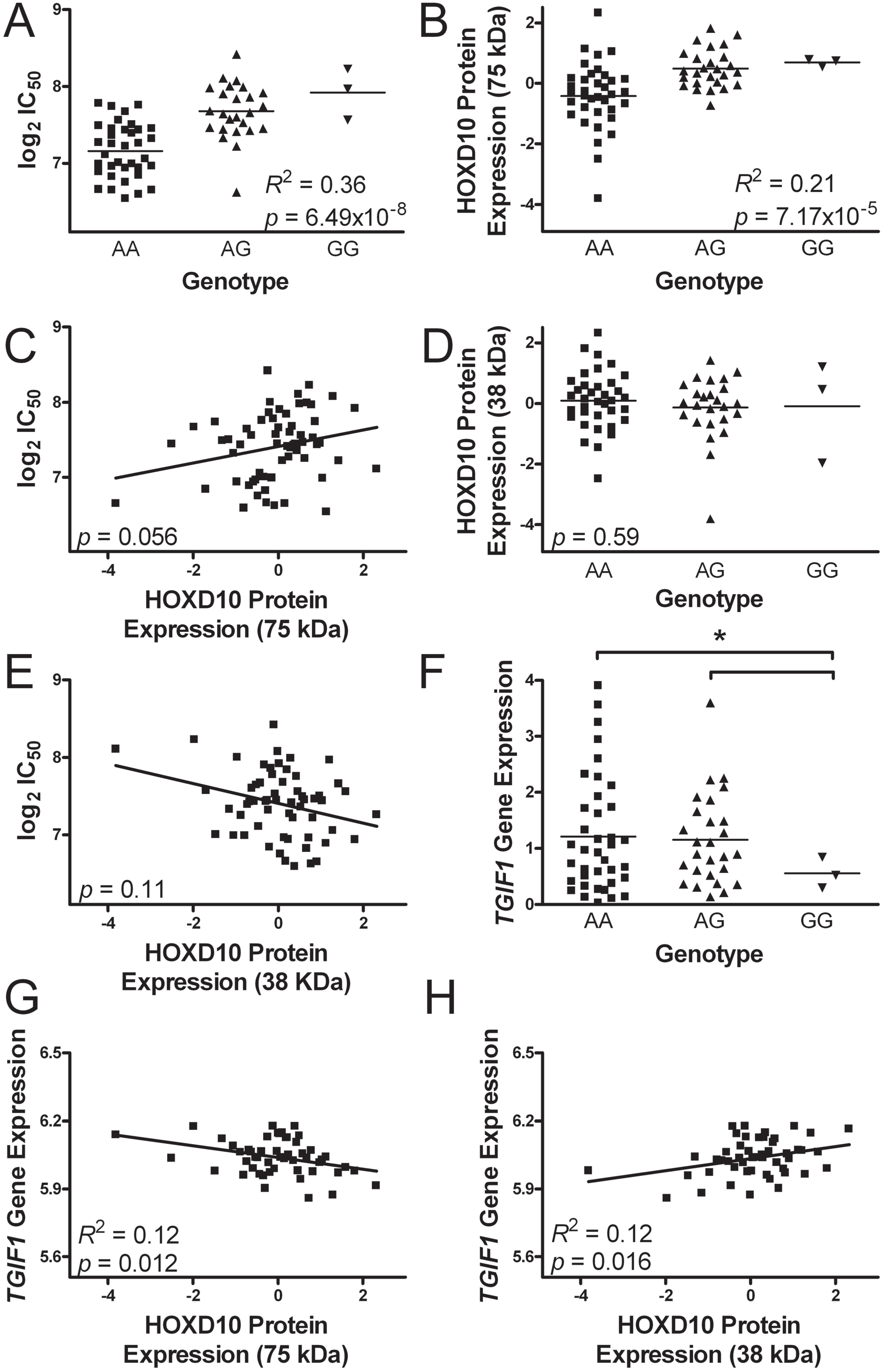
2.5. Evaluation of the Most Significant Single Nucleotide Polymorphism Associated with Colistin Cytotoxicity
2.6. chr18:3417240 and Homeobox Protein D10 Expression
2.7. Transforming Growth Factor β-Induced Factor-1 Gene Expression
2.8. Homeobox Protein D10 Expression Is Upregulated in the Kidney in Response to Colistin, Independent of Its Gene Expression
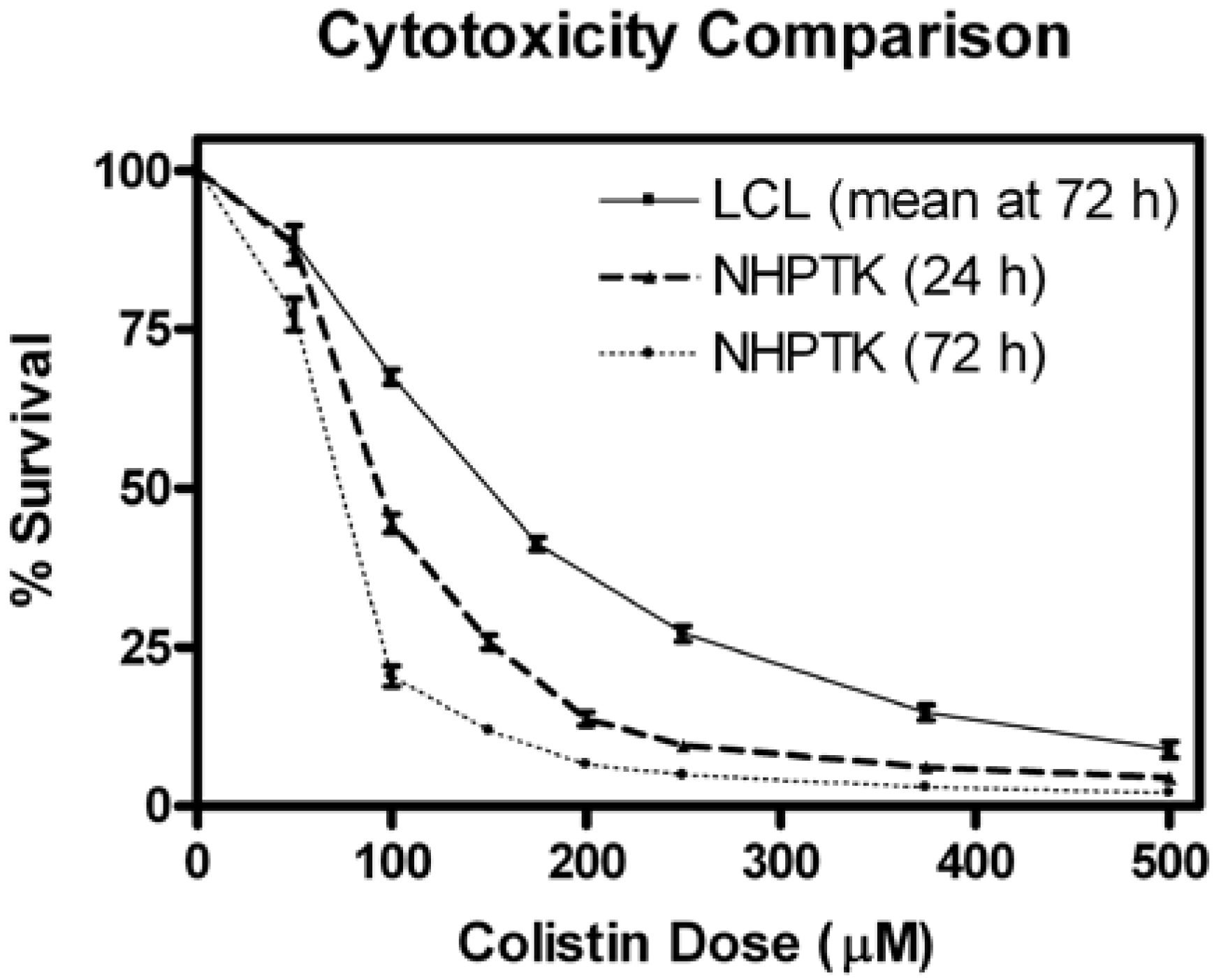
2.9. Colistin-Induced Cytotoxicity in Human Proximal Tubular Cells
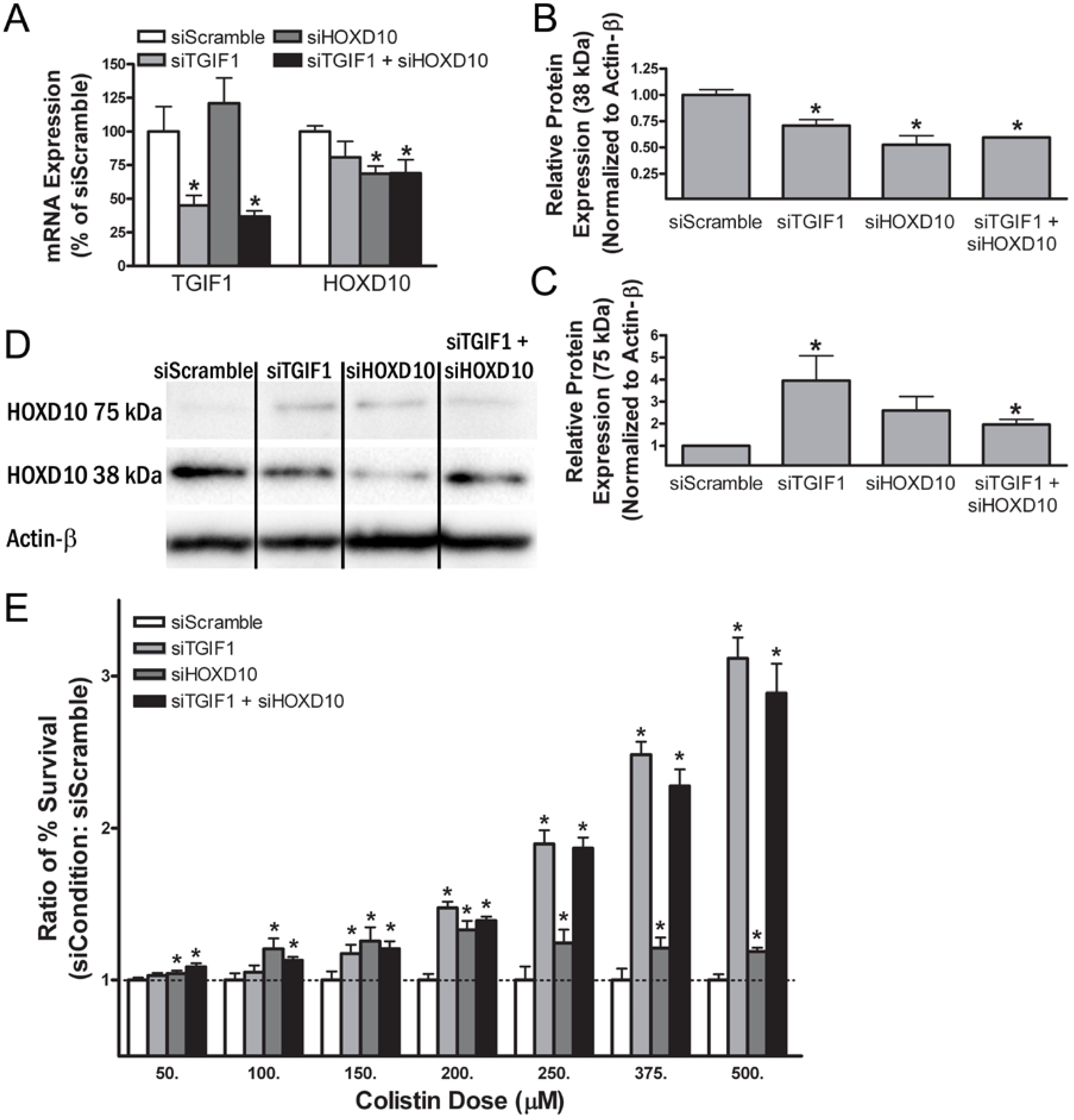
2.10. Smal Interfering RNA Knockdown of Transforming Growth Factor β-Induced Factor-1 and Homeobox Protein D10 Alters Gene and Protein Expression in Renal Cells
2.11. Small Interfering RNA Knockdown of Transforming Growth Factor β-Induced Factor-1 and Homeobox Protein D10 Alters Sensitivity to Colistin in Normal Human Proximal Tubular Kidney Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Drug
4.2. Colistin-Induced Cytotoxicity
4.3. Genome-Wide Association and Gene Expression Association with Phenotype
4.4. Expression Quantittative Trait Loci and Protein Quantittative Trait Loci Enrichment Analysis
4.5. Animal Studies and Immunohistochemistry
4.6. Small Interfering RNA Nucleofection
4.7. Quantitative Quantitative Real-Time Polymerase Chain Reaction
4.8. Protein Quantitation by Immunoblot in Normal Human Proximal Tubular Kidney Cells
4.9. Drug Treatment after Nucleofection
4.10. Small Interfering RNA Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, J.; Nation, R.L.; Milne, R.W.; Turnidge, J.D.; Coulthard, K. Evaluation of colistin as an agent against multi-resistant gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; de Santis, E.R.; Kuper, J. Resurgence of colistin use. Am. J. Health Syst. Pharm. 2007, 64, 2462–2466. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Azad, M.A.; Akter, J.; Rogers, K.L.; Nation, R.L.; Velkov, T.; Li, J. Major pathways of polymyxin-induced apoptosis in rat kidney proximal tubular cells. Antimicrob. Agents Chemother. 2015, 59, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Li, J.; Li, J. New insight in colistin induced neurotoxicity with the mitochondrial dysfunction in mice central nervous tissues. Exp. Toxicol. Pathol. 2013, 65, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Deryke, C.A.; Crawford, A.J.; Uddin, N.; Wallace, M.R. Colistin dosing and nephrotoxicity in a large community teaching hospital. Antimicrob. Agents Chemother. 2010, 54, 4503–4505. [Google Scholar] [CrossRef] [PubMed]
- Hartzell, J.D.; Neff, R.; Ake, J.; Howard, R.; Olson, S.; Paolino, K.; Vishnepolsky, M.; Weintrob, A.; Wortmann, G. Nephrotoxicity associated with intravenous colistin (colistimethate sodium) treatment at a tertiary care medical center. Clin. Infect. Dis. 2009, 48, 1724–1728. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Jeon, M.; Choo, E.; Lee, E.; Kim, T.; Jun, J.B.; Gil, H.W. Early acute kidney injury is a risk factor that predicts mortality in patients treated with colistin. Nephron Clin. Pract. 2011, 117, c284–c288. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Bishara, J.; Levcovich, A.; Chowers, M.; Goldberg, E.; Singer, P.; Lev, S.; Leon, P.; Raskin, M.; Yahav, D.; et al. Effectiveness and safety of colistin: Prospective comparative cohort study. J. Antimicrob. Chemother. 2010, 65, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, C.; Mykietiuk, A.; Temporiti, E.; Stryjewski, M.E.; Herrera, F.; Bonvehi, P. Nephrotoxicity associated with the use of intravenous colistin. Scand. J. Infect. Dis. 2009, 41, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, K.H.; Yoo, S.; Pai, H. Clinical characteristics and risk factors of colistin-induced nephrotoxicity. Int. J. Antimicrob. Agents 2009, 34, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.A.; Lee, J.E.; Huh, W.; Peck, K.R.; Kim, Y.G.; Kim, D.J.; Oh, H.Y. Predictors of acute kidney injury associated with intravenous colistin treatment. Int. J. Antimicrob. Agents 2010, 35, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Eadon, M.T.; Wheeler, H.E.; Stark, A.L.; Zhang, X.; Moen, E.L.; Delaney, S.M.; Im, H.K.; Cunningham, P.N.; Zhang, W.; Dolan, M.E. Genetic and epigenetic variants contributing to clofarabine cytotoxicity. Hum. Mol. Genet. 2013, 22, 4007–4020. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, H.E.; Dolan, M.E. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics 2012, 13, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Gamazon, E.R.; Wheeler, H.E.; Dolan, M.E. Clinical translation of cell-based pharmacogenomic discovery. Clin. Pharmacol. Ther. 2012, 92, 425–427. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.J.; Kraja, A.T.; Lin, S.J.; Yen-Revollo, J.L.; Marsh, S.; Province, M.A.; McLeod, H.L. Association of thymidylate synthase variants with 5-fluorouracil cytotoxicity. Pharmacogenet. Genom. 2009, 19, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Schaid, D.J.; Abo, R.P.; Kalari, K.; Fridley, B.L.; Feng, Q.; Jenkins, G.; Batzler, A.; Brisbin, A.G.; Cunningham, J.M.; et al. Genetic association with overall survival of taxane-treated lung cancer patients—A genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer 2012, 12, 422. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.C.; Havener, T.M.; Medina, M.W.; Auman, J.T.; Mangravite, L.M.; Krauss, R.M.; McLeod, H.L.; Motsinger-Reif, A.A. A genome-wide association analysis of temozolomide response using lymphoblastoid cell lines shows a clinically relevant association with mgmt. Pharmacogenet. Genomics 2012, 22, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Bolotin, E.; Theusch, E.; Huang, H.; Medina, M.W.; Krauss, R.M. Prediction of LDL cholesterol response to statin using transcriptomic and genetic variation. Genome Biol. 2014, 15, 460. [Google Scholar] [CrossRef] [PubMed]
- Mangravite, L.M.; Engelhardt, B.E.; Medina, M.W.; Smith, J.D.; Brown, C.D.; Chasman, D.I.; Mecham, B.H.; Howie, B.; Shim, H.; Naidoo, D.; et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 2013, 502, 377–380. [Google Scholar] [CrossRef] [PubMed]
- International HapMap Consortium. The international hapmap project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Duan, S.; Kistner, E.O.; Bleibel, W.K.; Huang, R.S.; Clark, T.A.; Chen, T.X.; Schweitzer, A.C.; Blume, J.E.; Cox, N.J.; et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008, 82, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Hause, R.J.; Stark, A.L.; Antao, N.N.; Gorsic, L.K.; Chung, S.H.; Brown, C.D.; Wong, S.S.; Gill, D.F.; Myers, J.L.; To, L.A.; et al. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. Am. J. Hum. Genet. 2014, 95, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Crews, K.R.; Pounds, S.; Cao, X.; Feldberg, T.; Ghodke, Y.; Gandhi, V.; Plunkett, W.; Dolan, M.E.; Hartford, C.; et al. Genetic variants in cytosolic 5’-nucleotidase II are associated with its expression and cytarabine sensitivity in hapmap cell lines and in patients with acute myeloid leukemia. J. Pharmacol. Exp. Ther. 2011, 339, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Ziliak, D.; O’Donnell, P.H.; Im, H.K.; Gamazon, E.R.; Chen, P.; Delaney, S.; Shukla, S.; Das, S.; Cox, N.J.; Vokes, E.E.; et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl. Res. 2011, 157, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Yang, W.; Fan, Y.; Stocco, G.; Crews, K.R.; Yang, J.J.; Paugh, S.W.; Pui, C.H.; Evans, W.E.; Relling, M.V. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia 2011, 25, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.L.; Moyer, A.M.; Fridley, B.L.; Schaid, D.J.; Niu, N.; Batzler, A.J.; Jenkins, G.D.; Abo, R.P.; Li, L.; Cunningham, J.M.; et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin. Cancer Res. 2011, 17, 5801–5811. [Google Scholar] [CrossRef] [PubMed]
- Grundberg, E.; Small, K.S.; Hedman, A.K.; Nica, A.C.; Buil, A.; Keildson, S.; Bell, J.T.; Yang, T.P.; Meduri, E.; Barrett, A.; et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012, 44, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Goring, H.H. Tissue specificity of genetic regulation of gene expression. Nat. Genet. 2012, 44, 1077–1078. [Google Scholar] [CrossRef] [PubMed]
- Travis, L.B.; Fossa, S.D.; Sesso, H.D.; Frisina, R.D.; Herrmann, D.N.; Beard, C.J.; Feldman, D.R.; Pagliaro, L.C.; Miller, R.C.; Vaughn, D.J.; et al. Chemotherapy-induced peripheral neurotoxicity and ototoxicity: New paradigms for translational genomics. J. Natl. Cancer Inst. 2014. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, H.E.; Gonzalez-Neira, A.; Pita, G.; de la Torre-Montero, J.C.; Alonso, R.; Lopez-Fernandez, L.A.; Alba, E.; Martin, M.; Dolan, M.E. Identification of genetic variants associated with capecitabine-induced hand-foot syndrome through integration of patient and cell line genomic analyses. Pharmacogenet. Genom. 2014, 24, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, H.E.; Gamazon, E.R.; Wing, C.; Njiaju, U.O.; Njoku, C.; Baldwin, R.M.; Owzar, K.; Jiang, C.; Watson, D.; Shterev, I.; et al. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of paclitaxel-induced sensory peripheral neuropathy. Clin. Cancer Res. 2013, 19, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Hneino, M.; Francois, A.; Buard, V.; Tarlet, G.; Abderrahmani, R.; Blirando, K.; Hoodless, P.A.; Benderitter, M.; Milliat, F. The TGF-β/Smad repressor TG-interacting factor 1 (TGIF1) plays a role in radiation-induced intestinal injury independently of a smad signaling pathway. PLoS ONE 2012, 7, e35672. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Peng, S.; Sun, Z.; Li, H.; Yang, R. Curcumin suppresses TGF-β signaling by inhibition of tgif degradation in scleroderma fibroblasts. Biochem. Biophys. Res. Commun. 2011, 411, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Herbert, B.S.; Grimes, B.R.; Xu, W.M.; Werner, M.; Ward, C.; Rossetti, S.; Harris, P.; Bello-Reuss, E.; Ward, H.H.; Miller, C.; et al. A telomerase immortalized human proximal tubule cell line with a truncation mutation (Q4004X) in polycystin-1. PLoS ONE 2013, 8, e55191. [Google Scholar] [CrossRef] [PubMed]
- Eadon, M.T.; Hack, B.K.; Alexander, J.J.; Xu, C.; Dolan, M.E.; Cunningham, P.N. Cell cycle arrest in a model of colistin nephrotoxicity. Physiol. Genom. 2013, 45, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Huang, R.S.; Cox, N.J.; Dolan, M.E. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 2010, 107, 9287–9292. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Marioni, J.C.; Pai, A.A.; Degner, J.F.; Engelhardt, B.E.; Nkadori, E.; Veyrieras, J.B.; Stephens, M.; Gilad, Y.; Pritchard, J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 2010, 464, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.L.; Hause, R.J., Jr.; Gorsic, L.K.; Antao, N.N.; Wong, S.S.; Chung, S.H.; Gill, D.F.; Im, H.K.; Myers, J.L.; White, K.P.; et al. Protein quantitative trait loci identify novel candidates modulating cellular response to chemotherapy. PLoS Genet. 2014, 10, e1004192. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, K.; Green, N.C.; Rambaldi, I.; Saragovi, H.U.; Featherstone, M.S. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 1999, 19, 7577–7588. [Google Scholar] [CrossRef] [PubMed]
- Phelan, M.L.; Featherstone, M.S. Distinct HOX N-terminal arm residues are responsible for specificity of DNA recognition by hox monomers and HOX·PBX heterodimers. J. Biol. Chem. 1997, 272, 8635–8643. [Google Scholar] [CrossRef] [PubMed]
- Stranger, B.E.; Nica, A.C.; Forrest, M.S.; Dimas, A.; Bird, C.P.; Beazley, C.; Ingle, C.E.; Dunning, M.; Flicek, P.; Koller, D.; et al. Population genomics of human gene expression. Nat. Genet. 2007, 39, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Wieser, M.; Stadler, G.; Jennings, P.; Streubel, B.; Pfaller, W.; Ambros, P.; Riedl, C.; Katinger, H.; Grillari, J.; Grillari-Voglauer, R. Htert alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 2008, 295, F1365–F1375. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ogawa, K.; Nagarajan, R.P.; Zhang, M.; Kuang, C.; Chen, Y. Regulation of TG-interacting factor by transforming growth factor-β. Biochem. J. 2003, 371, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Pessah, M.; Prunier, C.; Marais, J.; Ferrand, N.; Mazars, A.; Lallemand, F.; Gauthier, J.M.; Atfi, A. c-Jun interacts with the corepressor TG-interacting factor (TGIF) to suppress Smad2 transcriptional activity. Proc. Natl. Acad. Sci. USA 2001, 98, 6198–6203. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Womack, B.; Wotton, D.; Guo, Y.; Shyr, Y.; Dave, U.; Li, C.; Hiebert, S.; Brandt, S.; Hamid, R. TGIF1 regulates quiescence and self-renewal of hematopoietic stem cells. Mol. Cell. Biol. 2013, 33, 4824–4833. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.; Patterson, J.; Brandt, S.J. Genomic structure, alternative splicing and expression of TG-interacting factor, in human myeloid leukemia blasts and cell lines. Biochim. Biophys. Acta 2008, 1779, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Liborio, T.N.; Ferreira, E.N.; Aquino Xavier, F.C.; Carraro, D.M.; Kowalski, L.P.; Soares, F.A.; Nunes, F.D. TGIF1 splicing variant 8 is overexpressed in oral squamous cell carcinoma and is related to pathologic and clinical behavior. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 116, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Gabellini, D.; Colaluca, I.N.; Vodermaier, H.C.; Biamonti, G.; Giacca, M.; Falaschi, A.; Riva, S.; Peverali, F.A. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003, 22, 3715–3724. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, C.C.; Der-Avakian, A.; Spyropoulos, D.D.; Tieu, D.D.; Carpenter, E.M. Targeted disruption of HOXD9 and HOXD10 alters locomotor behavior, vertebral identity, and peripheral nervous system development. Dev. Biol. 1999, 216, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, F.; Zhao, D.Y.; Zhang, J.S.; Lv, Y.; Li-Ling, J. Interaction between TBX1 and HOXD10 and connection with TGFβ-BMP signal pathway during kidney development. Gene 2014, 536, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Redline, R.W.; Hudock, P.; MacFee, M.; Patterson, P. Expression of abdb-type homeobox genes in human tumors. Lab. Investig. 1994, 71, 663–670. [Google Scholar] [PubMed]
- Reddy, S.D.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008, 68, 8195–8200. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nie, S.; Chang, C.; Qiu, T.; Cao, X. Smads oppose HOX transcriptional activities. Exp. Cell Res. 2006, 312, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Eadon, M.T.; Desta, Z.; Levy, K.D.; Decker, B.S.; Pierson, R.C.; Pratt, V.M.; Callaghan, J.T.; Rosenman, M.B.; Carpenter, J.S.; Holmes, A.M.; et al. Implementation of a pharmacogenomics consult service to support the ingenious trial. Clin. Pharmacol. Ther. 2016, 100, 63–66. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, P.H.; Danahey, K.; Jacobs, M.; Wadhwa, N.R.; Yuen, S.; Bush, A.; Sacro, Y.; Sorrentino, M.J.; Siegler, M.; Harper, W.; et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care—Initial results of the university of chicago “1200 patients project”. Am. J. Med. Genet. Part C Semin. Med. Genet. 2014, 166C, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Levy, K.D.; Decker, B.S.; Carpenter, J.S.; Flockhart, D.A.; Dexter, P.R.; Desta, Z.; Skaar, T.C. Prerequisites to implementing a pharmacogenomics program in a large health-care system. Clin. Pharmacol. Ther. 2014, 96, 307–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Milne, R.W.; Nation, R.L.; Turnidge, J.D.; Coulthard, K. Stability of colistin and colistin methanesulfonate in aqueous media and plasma as determined by high-performance liquid chromatography. Antimicrob. Agents Chemother. 2003, 47, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Vaara, T. The novel polymyxin derivative NAB739 is remarkably less cytotoxic than polymyxin B and colistin to human kidney proximal tubular cells. Int. J. Antimicrob. Agents 2013, 41, 292–293. [Google Scholar] [CrossRef] [PubMed]
- Keirstead, N.D.; Wagoner, M.P.; Bentley, P.; Blais, M.; Brown, C.; Cheatham, L.; Ciaccio, P.; Dragan, Y.; Ferguson, D.; Fikes, J.; et al. Early prediction of polymyxin-induced nephrotoxicity with next-generation urinary kidney injury biomarkers. Toxicol. Sci. 2013, 137, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Genomes Project Consortium; Abecasis, G.R.; Altshuler, D.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Gibbs, R.A.; Hurles, M.E.; McVean, G.A. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [Green Version]
- Genomes Project Consortium; Abecasis, G.R.; Auton, A.; Brooks, L.D.; de Pristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar]
- Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [Green Version]
- R Core Team (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available online: http://www.R-project.org/ (accessed on 15 January 2017).
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. Nih image to imagej: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eadon, M.T.; Hause, R.J.; Stark, A.L.; Cheng, Y.-H.; Wheeler, H.E.; Burgess, K.S.; Benson, E.A.; Cunningham, P.N.; Bacallao, R.L.; Dagher, P.C.; et al. Genetic Variants Contributing to Colistin Cytotoxicity: Identification of TGIF1 and HOXD10 Using a Population Genomics Approach. Int. J. Mol. Sci. 2017, 18, 661. https://doi.org/10.3390/ijms18030661
Eadon MT, Hause RJ, Stark AL, Cheng Y-H, Wheeler HE, Burgess KS, Benson EA, Cunningham PN, Bacallao RL, Dagher PC, et al. Genetic Variants Contributing to Colistin Cytotoxicity: Identification of TGIF1 and HOXD10 Using a Population Genomics Approach. International Journal of Molecular Sciences. 2017; 18(3):661. https://doi.org/10.3390/ijms18030661
Chicago/Turabian StyleEadon, Michael T., Ronald J. Hause, Amy L. Stark, Ying-Hua Cheng, Heather E. Wheeler, Kimberly S. Burgess, Eric A. Benson, Patrick N. Cunningham, Robert L. Bacallao, Pierre C. Dagher, and et al. 2017. "Genetic Variants Contributing to Colistin Cytotoxicity: Identification of TGIF1 and HOXD10 Using a Population Genomics Approach" International Journal of Molecular Sciences 18, no. 3: 661. https://doi.org/10.3390/ijms18030661





