CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis
Abstract
:1. Introduction
2. Results
2.1. Isolation and Phylogenetic Analysis of the CmMYB19 cDNA Sequence
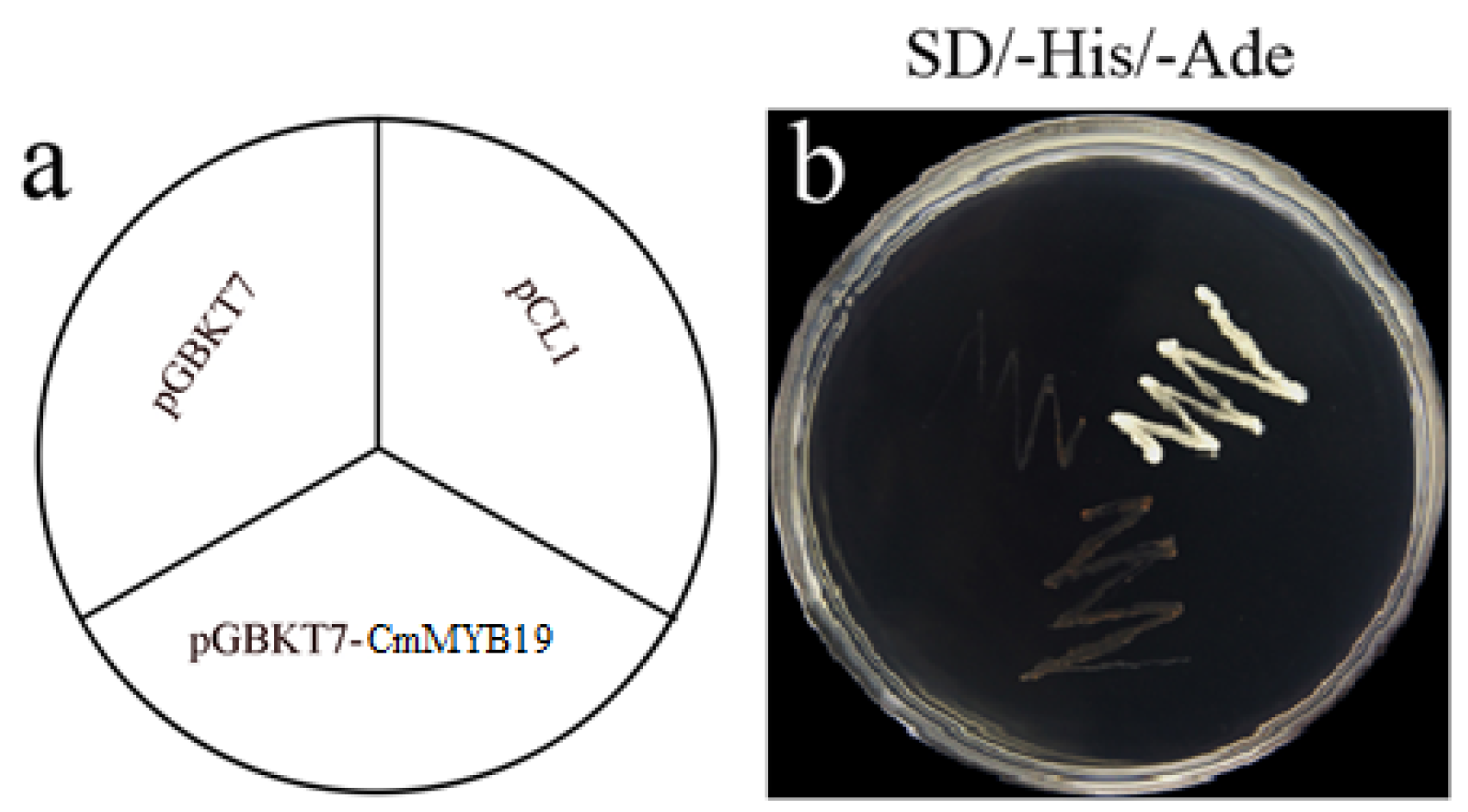
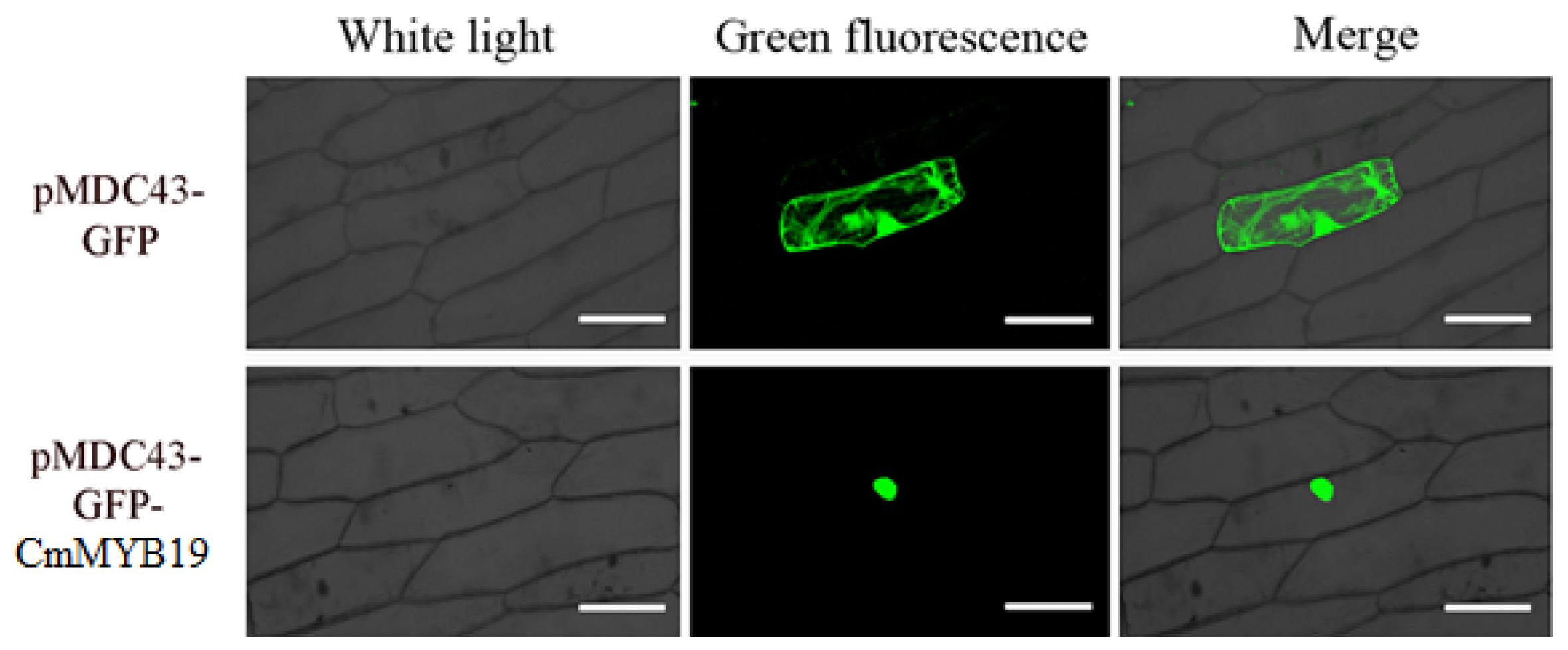
2.2. Transcriptional Activation and Sub-Cellular Localization
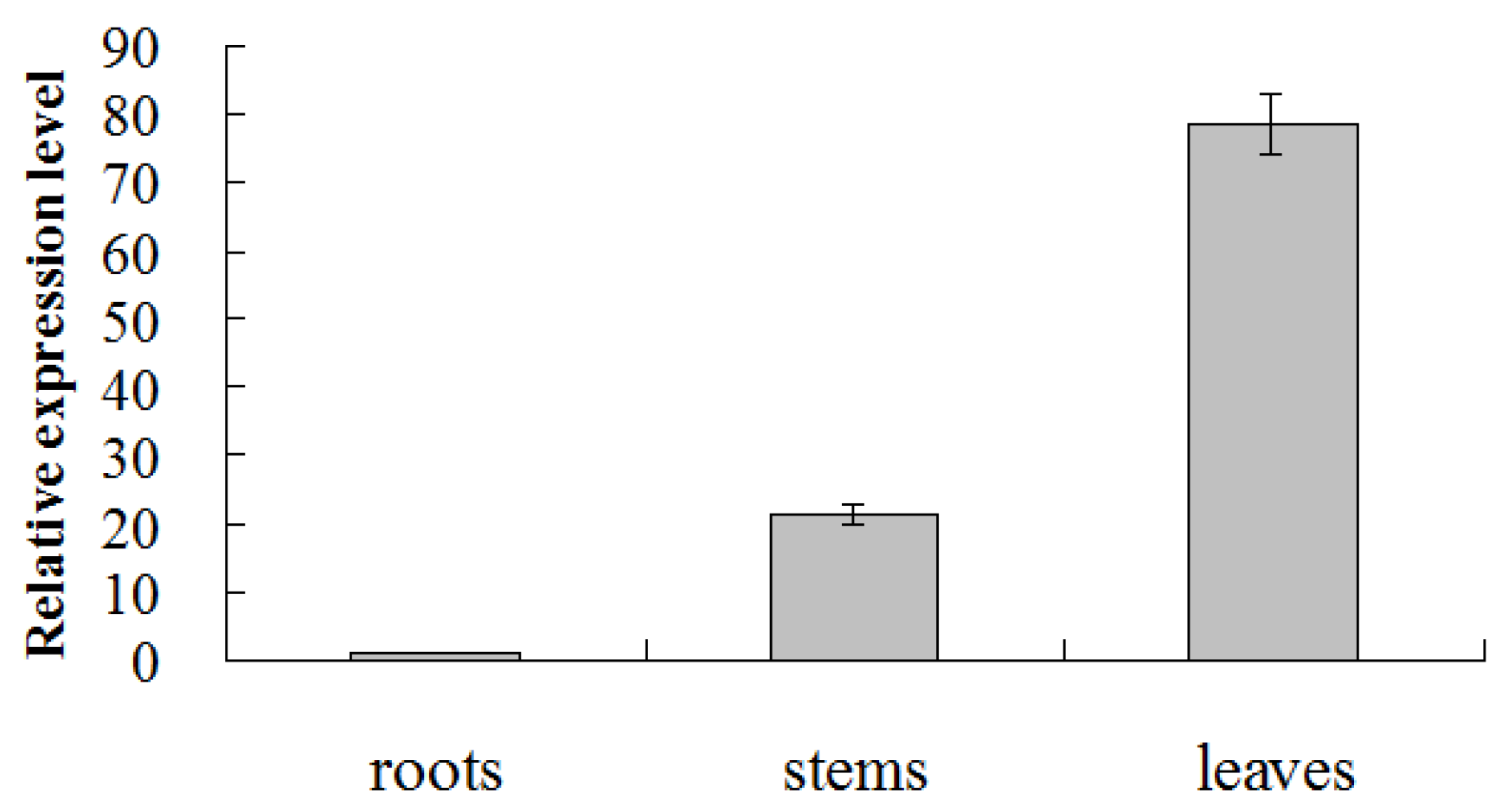
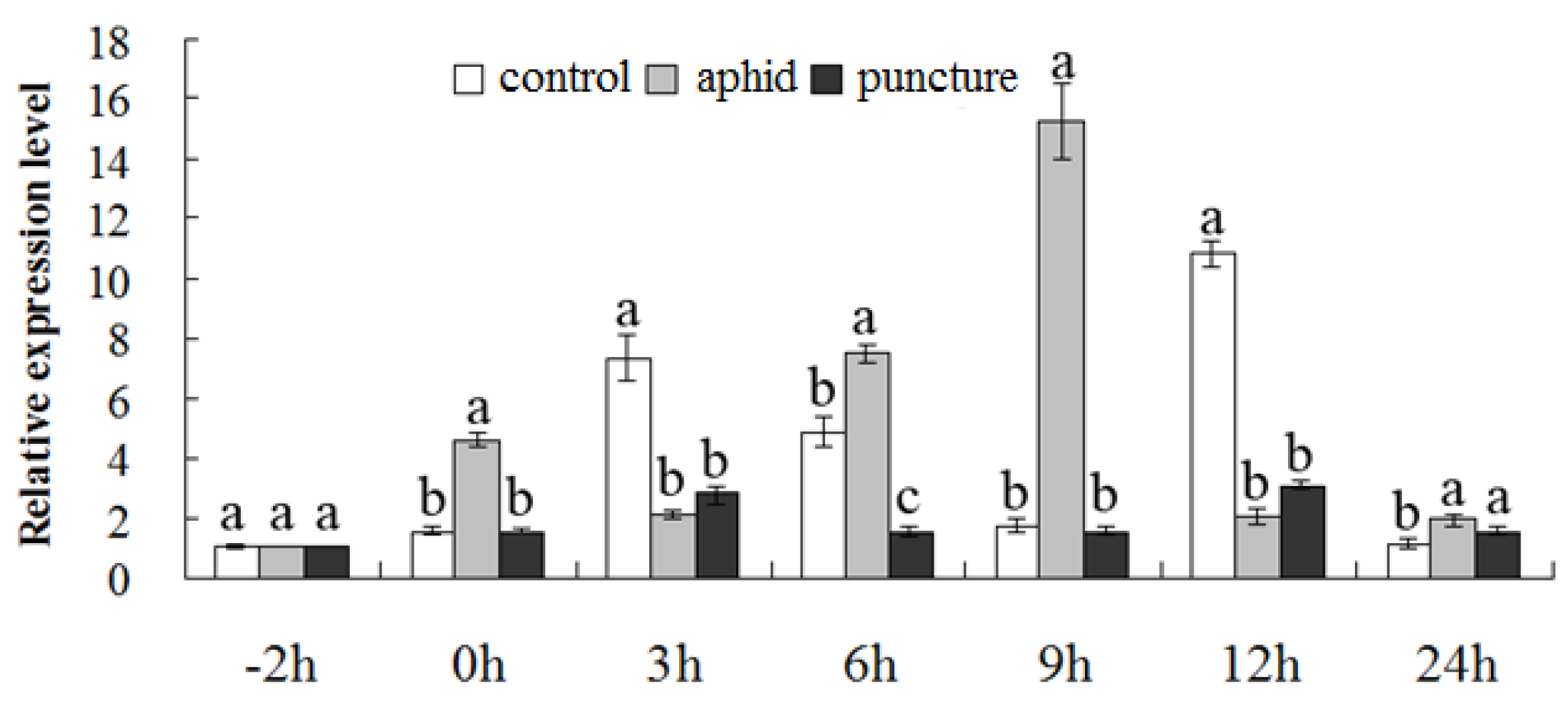
2.3. Transcription of CmMYB19 in the Chrysanthemum Plant
2.4. CmMYB19 Over-Expression Enhanced Aphid Tolerance
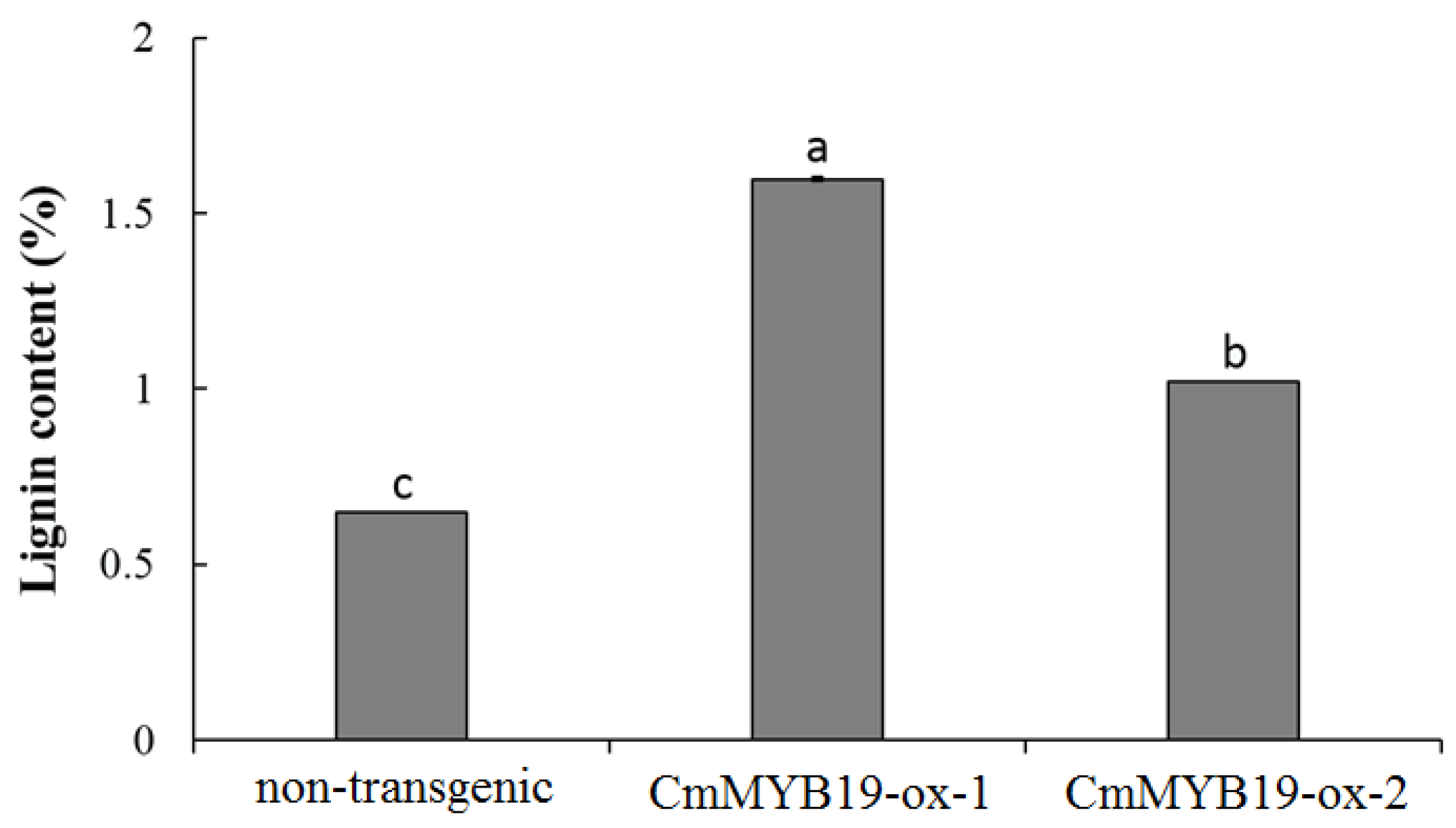
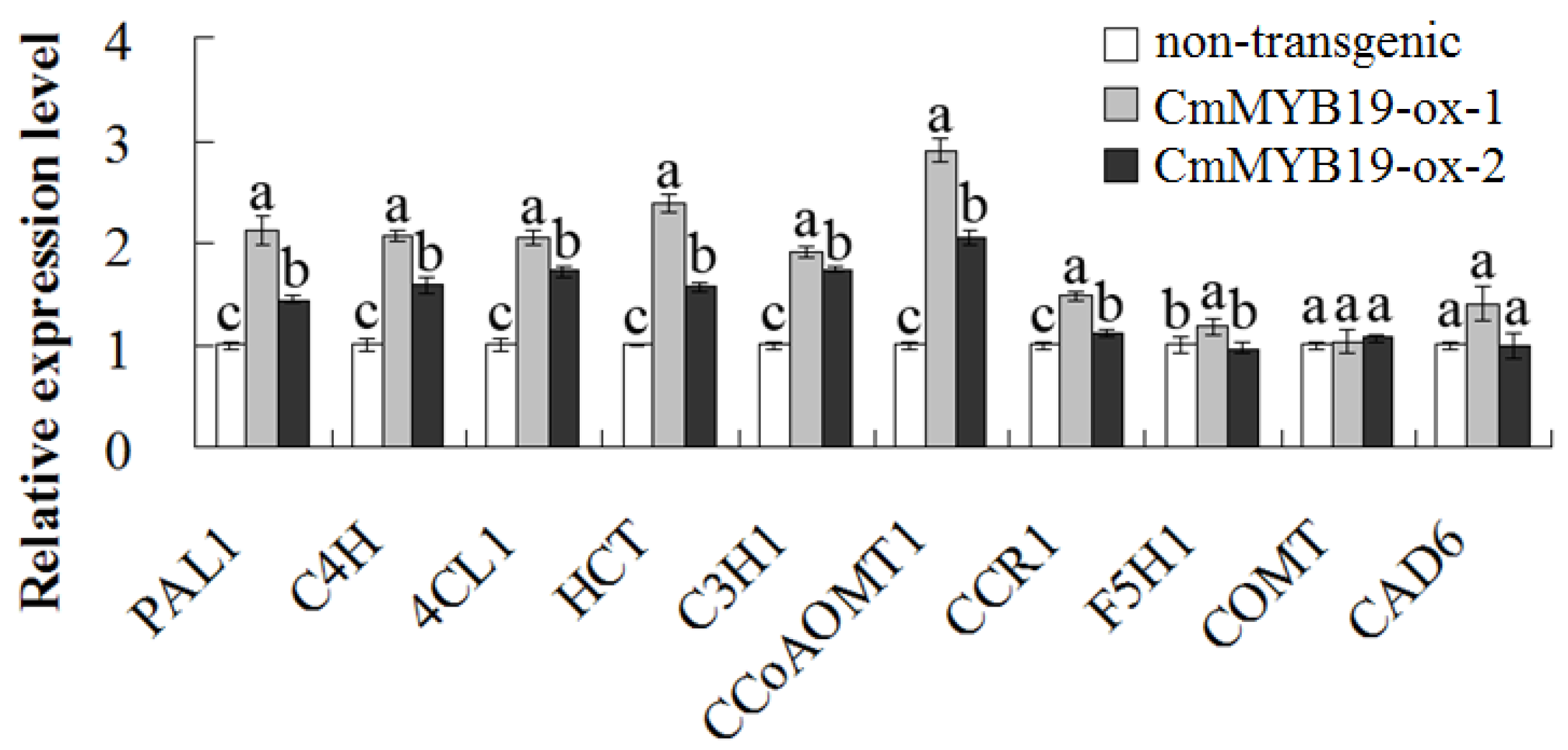
2.5. Lignin Content and the Transcript Abundance of Genes Involved in Lignin Synthesis in CmMYB19 Over-Expressors
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growing Conditions
4.2. Isolation and Sequence Analysis of CmMYB19
4.3. Transcriptional Activation Assay and Sub-Cellular Localization of CmMYB19
4.4. Transcription of CmMYB19 and Its Response to Aphid Infestation
4.5. Generation of Transgenic Chrysanthemum and the Determination of Aphid Tolerance
4.6. Determination of Lignin Content and the Transcription of Lignin Synthesis Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.-F.; Yang, W.-J.; Wu, Y.-M.; Huang, Y.-B.; Tang, Y.-X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Wang, X.; Li, C.; Lu, W.; Yang, L.; Jiang, Y.; Luo, K. Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS ONE 2013, 8, e76369. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhuang, Y.; Qi, G.; Wang, D.; Liu, H.; Wang, K.; Chai, G.; Zhou, G. Poplar PdMYB221 is involved in the direct and indirect regulation of secondary wall biosynthesis during wood formation. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, S.; Lu, A.; Chen, F.; Tang, F.; Guan, Z.; Teng, N. Production and characterisation of the intergeneric hybrids between Dendranthema morifolium and Artemisia vulgaris exhibiting enhanced resistance to chrysanthemum aphid (Macrosiphoniella sanbourni). Planta 2010, 231, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Goggin, F.L. Plant–aphid interactions: Molecular and ecological perspectives. Curr. Opin. Plant Biol. 2007, 10, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Yang, Q.; Pan, L.; Chi, X.; Chen, M.; Hu, D.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene 2014, 533, 332–345. [Google Scholar] [CrossRef] [PubMed]
- De Vos, M.; Denekamp, M.; Dicke, M.; Vuylsteke, M.; Van Loon, L.; Smeekens, S.C.; Pieterse, C. The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal. Behav. 2006, 1, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Lü, B.B.; Li, X.J.; Sun, W.W.; Li, L.; Gao, R.; Zhu, Q.; Tian, S.M.; Fu, M.Q.; Yu, H.L.; Tang, X.M. AtMYB44 regulates resistance to the green peach aphid and diamondback moth by activating EIN2-affected defences in Arabidopsis. Plant Biol. 2013, 15, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Shan, H.; Chen, S.; Jiang, J.; Gu, C.; Zhou, G.; Chen, Y.; Song, A.; Chen, F. The heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e65680. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H. Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lü, B.; Wang, X.; Zhang, C.; Zhang, S.; Qian, J.; Chen, L.; Shi, H.; Dong, H. Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. J. Biosci. 2010, 35, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Kondo, M.; Aya, K.; Miyao, A.; Sato, Y.; Antonio, B.A.; Namiki, N.; Nagamura, Y.; Matsuoka, M. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013, 54, 1791–1802. [Google Scholar] [CrossRef] [PubMed]
- Öhman, D.; Demedts, B.; Kumar, M.; Gerber, L.; Gorzsás, A.; Goeminne, G.; Hedenström, M.; Ellis, B.; Boerjan, W.; Sundberg, B. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 2013, 73, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xu, Z.; Song, J.; Conner, K.; Barrena, G.V.; Wilson, Z.A. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 2007, 19, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Zou, W.; Feng, Y.; Zhang, M.; Zhang, J.; Tu, F.; Xie, G.; Wang, L.; Wang, Y.; Klie, S. An integrated genomic and metabolomic framework for cell wall biology in rice. BMC Genom. 2014, 15, 596. [Google Scholar] [CrossRef] [PubMed]
- Chantreau, M.; Portelette, A.; Dauwe, R.; Kiyoto, S.; Crônier, D.; Morreel, K.; Arribat, S.; Neutelings, G.; Chabi, M.; Boerjan, W. Ectopic lignification in the flax lignified bast fiber1 mutant stem is associated with tissue-specific modifications in gene expression and cell wall composition. Plant Cell 2014, 26, 4462–4482. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, F.; Chen, S.; Lv, G.; Deng, Y.; Fang, W.; Liu, Z.; Guan, Z.; He, C. Chrysanthemum leaf epidermal surface morphology and antioxidant and defense enzyme activity in response to aphid infestation. J. Plant Physiol. 2011, 168, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, Y.; Peng, X.; Wang, Q.; Harris, M.; Ge, F. Up-regulation of abscisic acid signaling pathway facilitates aphid xylem absorption and osmoregulation under drought stress. J. Exp. Bot. 2016, 67, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, E.; Porth, I.; Chen, J.-G.; Mansfield, S.D.; Douglas, C.J. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Rahantamalala, A.; Rech, P.; Martinez, Y.; Chaubet-Gigot, N.; Grima-Pettenati, J.; Pacquit, V. Coordinated transcriptional regulation of two key genes in the lignin branch pathway-CAD and CCR-is mediated through MYB-binding sites. BMC Plant Biol. 2010, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yin, X.-R.; Zeng, J.-K.; Ge, H.; Song, M.; Xu, C.-J.; Li, X.; Ferguson, I.B.; Chen, K.-S. Activator-and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Winzell, A.; Aspeborg, H.; Wang, Y.; Ezcurra, I. Conserved CA-rich motifs in gene promoters of Pt × tMYB021-responsive secondary cell wall carbohydrate-active enzymes in Populus. Biochem. Biophys. Res. Commun. 2010, 394, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Jond, C.; Debeaujon, I.; Caboche, M.; Lepiniec, L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 2001, 13, 2099–2114. [Google Scholar] [PubMed]
- Xia, X.; Shao, Y.; Jiang, J.; Ren, L.; Chen, F.; Fang, W.; Guan, Z.; Chen, S. Gene expression profiles responses to aphid feeding in chrysanthemum (Chrysanthemum morifolium). BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, Z.; Liu, T.; Wang, Y. Biodiversity, Phylogeny, and Antifungal Functions of Endophytic Fungi Associated with Zanthoxylum bungeanum. Int. J. Mol. Sci. 2016, 17, 1541. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Song, A.; Zhu, X.; Chen, F.; Jiang, J.; Chen, Y.; Sun, Y.; Shan, H.; Gu, C.; Li, P. The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 2012, 235, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Jiang, J.; Chen, S.; Fang, W.; Zhang, F.; Chen, F. The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 2015, 95, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Song, A.; Xin, J.; Chen, S.; Jiang, J.; Wang, Y.; Li, X.; Chen, F. CmWRKY15 facilitates Alternaria tenuissima infection of chrysanthemum. PLoS ONE 2015, 10, e0143349. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef] [PubMed]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chen, S.; Song, A.; Zhao, S.; Fang, W.; Guan, Z.; Liao, Y.; Jiang, J.; Chen, F. A putative high affinity phosphate transporter, CmPT1, enhances tolerance to Pi deficiency of chrysanthemum. BMC Plant Biol. 2014, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Fagerstedt, K.V.; Saranpää, P.; Tapanila, T.; Immanen, J.; Serra, J.A. A.; Nieminen, K. Determining the composition of lignins in different tissues of silver birch. Plants 2015, 4, 183–195. [Google Scholar] [CrossRef] [PubMed]



| Plants | MR * | IR | |
|---|---|---|---|
| Non-transgenic plants | 28.48 ± 0.262 a | 0 | |
| Transgenic plants | CmMYB19-ox-1 | 16.78 ± 0.548 c | 41.08 |
| CmMYB19-ox-2 | 19.68 ± 0.776 b | 30.90 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Sheng, L.; Zhang, H.; Du, X.; An, C.; Xia, X.; Chen, F.; Jiang, J.; Chen, S. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. Int. J. Mol. Sci. 2017, 18, 619. https://doi.org/10.3390/ijms18030619
Wang Y, Sheng L, Zhang H, Du X, An C, Xia X, Chen F, Jiang J, Chen S. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. International Journal of Molecular Sciences. 2017; 18(3):619. https://doi.org/10.3390/ijms18030619
Chicago/Turabian StyleWang, Yinjie, Liping Sheng, Huanru Zhang, Xinping Du, Cong An, Xiaolong Xia, Fadi Chen, Jiafu Jiang, and Sumei Chen. 2017. "CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis" International Journal of Molecular Sciences 18, no. 3: 619. https://doi.org/10.3390/ijms18030619
APA StyleWang, Y., Sheng, L., Zhang, H., Du, X., An, C., Xia, X., Chen, F., Jiang, J., & Chen, S. (2017). CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. International Journal of Molecular Sciences, 18(3), 619. https://doi.org/10.3390/ijms18030619








