The Role and Molecular Mechanism of Non-Coding RNAs in Pathological Cardiac Remodeling
Abstract
:1. Introduction
2. The Classification and Function of ncRNAs
3. miRNAs and Cardiac Remodeling
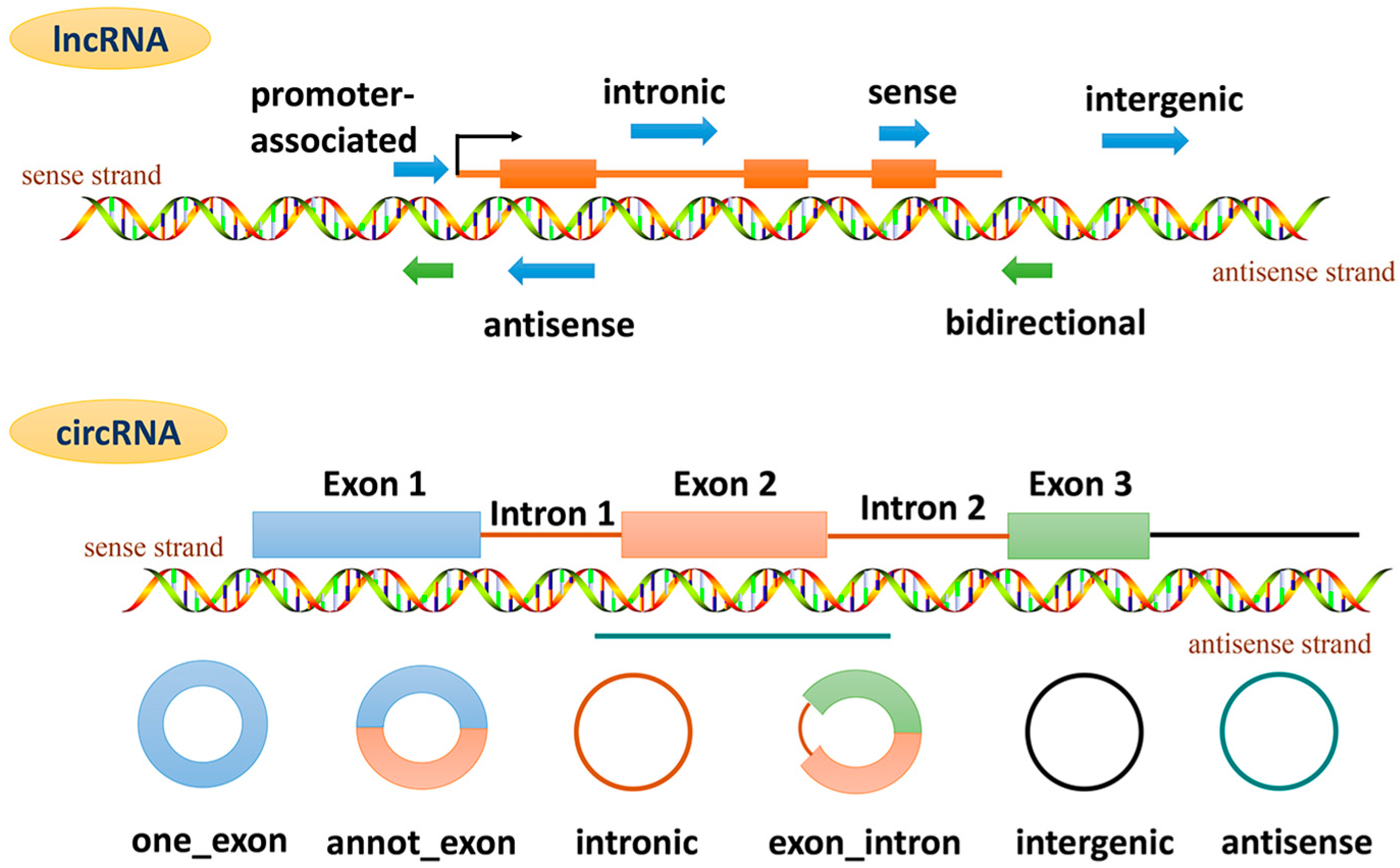
4. lncRNAs and Cardiac Remodeling
4.1. lncRNA and Epigenetic Regulation
4.1.1. lncRNA and Chromatin Remodeling
4.1.2. lncRNA and Histone Methylation
4.2. lncRNA as Molecular Sponge for miRNA
4.3. lncRNA Regulates the Expression of Adjacent Protein Coding Gene
4.4. lncRNA and Protein Complexes Bind to the Gene Promoter Region, Regulating Gene Expression
4.5. lncRNA as miRNA Precursor
4.6. lncRNA Modulates Protein Activity
4.7. Others
5. circRNAs and Cardiac Remodeling
5.1. circRNA Regulates the Adjacent Gene Expression
5.2. circRNA Functions as ceRNA
5.3. circRNA Binds to Specific Proteins, Altering Their Cellular Localization
6. Conclusions and Prospective
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Adcy6 | Adenylate cyclase 6 |
| ALDH2 | Aldehyde dehydrogenase 2 |
| Aldo | Aldosterone |
| AMPK | AMP-activated protein kinase |
| Ang II | Angiotensin II |
| ANT1 | Adenine nucleotide translocase 1 |
| APF | Autophagy promoting factor |
| A/R | Anoxia/reoxygenation |
| ASVD | Atherosclerotic vascular disease |
| Bcat2 | Branched chain amino acid transaminase 2 |
| BNP | B-type-natriuretic peptide |
| CAL | Coronary artery ligation |
| calcineurin-NFAT | Calcineurin-nuclear factor of activated T cells |
| CaMKIIδ | Ca2+/calmodulin-dependent protein kinase II δ-isoform |
| CARL | Cardiac apoptosis related lncRNA |
| Cdc42 | Cell division cycle 42 |
| Cdr1as | Antisense of cerebellar degeneration-related protein 1 |
| ceRNA | Competing endogenous RNA |
| Chaer | Cardiac-hypertrophy-associated epigenetic regulator |
| Chast | Cardiac hypertrophy-associated Transcript |
| CHRF | Cardiac hypertrophy related lncRNA |
| circRNAs | Circular RNAs |
| CnA | Calcineurin catalytic subunit |
| CTGF | Connective tissue growth factor |
| Dox | Doxorubicin |
| EZH2 | Zeste homolog 2 |
| FADD | Fas-Associated protein with Death Domain |
| Foxo3 | Forkhead box transcription factor 3 |
| Foxp1 | Forkhead box protein P1 |
| GATA4 | GATA Binding Protein 4 |
| GRB2 | Growth factor receptor-bound protein 2 |
| GWAS | Genome wide association studies |
| HADC4 | Histone Deacetylase 4 |
| hiPSC-CMs | Human induced pluripotent stem cell–derived cardiomyocytes |
| H2O2 | Hydrogen peroxide |
| HRCR | Heart-related circRNA |
| IGFR-1 | Insulin-like growth factor receptor 1 |
| IL | Interleukin |
| I/R | Ischemia-reperfusion |
| Iso | Isoprenaline |
| JAK-STAT | Janus Kinase- Signal transducers and activators of transcription |
| Jarid2 | Jumonji, AT rich interactive domain 2 |
| KSR1 | Kinase suppressor of ras 1 |
| lncRNAs | Long non-coding RNAs |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 |
| MAPKs | Mitogen-activated protein kinases |
| MFF | Mitochondrial fission factor |
| MI | Myocardial infarction |
| MIAT | Myocardial infarction associated transcript |
| miRNAs | MicroRNAs |
| MuRF1 | Muscle specific ring finger protein 1 |
| MyD88 | Myeloid differentiation factor88 |
| Myh7 | Myosin heavy chain 7 |
| ncRNAs | Non-coding RNAs |
| Nelf-A | Negative elongation factor complex member A |
| NF-κB | Nuclear factor-κB |
| NRF | Necrosis-related factor |
| PE | Phenylephrine |
| PHB2 | Prohibitin 2 |
| PI3K/Akt | Phosphatidylinositol 3-kinase/Akt |
| PIGF | Placental growth factor |
| Pink1 | PTEN Induced Putative Kinase 1 |
| piRISC | piRNA-induced silencing complex |
| piRNAs | Piwi-interacting RNAs |
| PKC | Protein kinase C |
| POFUT1 | Protein O-fucosyltranferase 1 |
| PPP1R10 | Protein Phosphatase 1 Regulatory Subunit 10 |
| PRC2 | Polycomb repressor complex 2 |
| PTEN | Phosphatase And Tensin Homolog |
| PURB | Purine-rich element binding protein B |
| Rab1a | Ras-related protein Rab-1A |
| RhoA | Ras homolog gene family, member A |
| RIPK | Receptor-interacting serine/threonine-protein kinase |
| ROS | Reactive oxygen species |
| rRNAs | Ribosomal RNAs |
| SEMA4B | Semaphorin 4B |
| siRNAs | Small interfering RNAs |
| Sirt4 | Sirtuin 4 |
| Skp2 | S-Phase Kinase-Associated Protein 2 |
| SMAD7 | SMAD Family Member 7 |
| snoRNAs | Small nucleolar RNA |
| SNPs | Single nucleotide polymorphisms |
| snRNAs | Small nuclear RNAs |
| Spry1 | Sprouty homolog 1 |
| TAC | Transverse aortic constriction |
| Tbx 5 | T-box 5 |
| Tg | Transgenic |
| tRNAs | Transfer RNAs |
| TNF-α | Tumor necrosis factor-α |
| UCA1 | Urothelial carcinoma-associated 1 |
| VEGF | Vascular endothelial growth factors |
| WHSC2 | Wolf-Hirschhorn Syndrome Candidate 2 |
| YAP | Yes-associated protein |
References
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grander, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Bi, C.; Clark, B.S.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes. Dev. 2006, 20, 1470–1484. [Google Scholar] [CrossRef] [PubMed]
- Beltran, M.; Puig, I.; Pena, C.; Garcia, J.M.; Alvarez, A.B.; Pena, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes. Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microrna sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Kang, J.Y.; Gou, L.T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.W.; Chen, R.; Fu, X.D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Londin, E.; Loher, P.; Telonis, A.G.; Quann, K.; Clark, P.; Jing, Y.; Hatzimichael, E.; Kirino, Y.; Honda, S.; Lally, M. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. USA 2015, 112, E1106–E1115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. MiR2disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Olson, E.N. Pervasive roles of micrornas in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Condorelli, G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Care, A.; Catalucci, D.; Felicetti, F.; Bonci, D.; Addario, A.; Gallo, P.; Bang, M.L.; Segnalini, P.; Gu, Y.; Dalton, N.D.; et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007, 13, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, F.; Luo, S.; Zhao, W.; Yang, T.; Zhang, G.; Gao, M.; Lu, R.; Shu, Y.; Mu, W.; et al. Let-7a is an antihypertrophic regulator in the heart via targeting calmodulin. Int. J. Biol. Sci. 2017, 13, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Diniz, G.P.; Lino, C.A.; Moreno, C.R.; Senger, N.; Barreto-Chaves, M.L. MicroRNA-1 overexpression blunts cardiomyocyte hypertrophy elicited by thyroid hormone. J. Cell. Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Zhou, J.; Li, P.F. miR-9 and NFATc3 regulate myocardin in cardiac hypertrophy. J. Biol. Chem. 2010, 285, 11903–11912. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhai, G.; Ji, Y.; Jing, H. MicroRNA-10a targets T-box 5 to inhibit the development of cardiac hypertrophy. Int. Heart J. 2017, 58, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.P.; Chen, J.; Seok, H.Y.; Zhang, Z.; Kataoka, M.; Hu, X.; Wang, D.Z. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 2013, 112, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Gurha, P.; Abreu-Goodger, C.; Wang, T.; Ramirez, M.O.; Drumond, A.L.; van Dongen, S.; Chen, Y.; Bartonicek, N.; Enright, A.J.; Lee, B.; et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation 2012, 125, 2751–2761. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Murtaza, I.; Wang, K.; Jiao, J.; Gao, J.; Li, P.F. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 12103–12108. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Z.; Xiao, W. microRNA-26a protects against cardiac hypertrophy via inhibiting GATA4 in rat model and cultured cardiomyocytes. Mol. Med. Rep. 2016, 14, 2860–2866. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Gao, X.M.; Winbanks, C.E.; Boey, E.J.; Tham, Y.K.; Kiriazis, H.; Gregorevic, P.; Obad, S.; Kauppinen, S.; Du, X.J.; et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 2012, 109, 17615–17620. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Yuan, M.; Zhou, R.; Bai, Q.; Zhang, W.; Zhang, M.; Huang, Y.; Shi, L. MicroRNA-101 inhibits rat cardiac hypertrophy by targeting Rab1a. J. Cardiovasc. Pharmacol. 2015, 65, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Chen, J.; Kataoka, M.; Huang, Z.P.; Ding, J.; Yan, J.; Hu, X.; Wang, D.Z. Loss of microRNA-155 protects the heart from pathological cardiac hypertrophy. Circ. Res. 2014, 114, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hwangbo, C.; Jaba, I.M.; Zhang, J.; Papangeli, I.; Han, J.; Mikush, N.; Larrivee, B.; Eichmann, A.; Chun, H.J.; et al. miR-182 modulates myocardial hypertrophic response induced by angiogenesis in heart. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Del Re, D.P.; Nakano, N.; Sciarretta, S.; Zhai, P.; Park, J.; Sayed, D.; Shirakabe, A.; Matsushima, S.; Park, Y.; et al. miR-206 mediates YAP-induced cardiac hypertrophy and survival. Circ. Res. 2015, 117, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Ucar, A.; Gupta, S.K.; Fiedler, J.; Erikci, E.; Kardasinski, M.; Batkai, S.; Dangwal, S.; Kumarswamy, R.; Bang, C.; Holzmann, A.; et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, Y.; Wang, X.; Li, R.; Yin, D. MicroRNA-365 accelerates cardiac hypertrophy by inhibiting autophagy via the modulation of Skp2 expression. Biochem. Biophys. Res. Commun. 2017, 484, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, J.; Ramanujam, D.; Sassi, Y.; Ahles, A.; Jentzsch, C.; Werfel, S.; Leierseder, S.; Loyer, X.; Giacca, M.; Zentilin, L.; et al. miR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation 2013, 127, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, X.; Fan, S.; Cui, G.; Shen, Z. MicroRNA-497 inhibits cardiac hypertrophy by targeting Sirt4. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, N.; Long, B.; Fan, Y.Y.; Liu, C.Y.; Zhou, Q.Y.; Murtaza, I.; Wang, K.; Li, P.F. Cardiac hypertrophy is negatively regulated by miR-541. Cell Death Dis. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Chaanine, A.H.; Kang, S.; Mukete, B.N.; Jeong, D.; Zhang, S.; Hajjar, R.J.; Lebeche, D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Schauerte, C.; Hubner, A.; Kolling, M.; Martino, F.; Scherf, K.; Batkai, S.; Zimmer, K.; Foinquinos, A.; Kaucsar, T.; et al. Osteopontin is indispensible for AP1-mediated angiotensin II-related miR-21 transcription during cardiac fibrosis. Eur. Heart J. 2015, 36, 2184–2196. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Cao, H.; Wang, Q.; Ye, J.; Sui, L.; Feng, J.; Cai, X.; Song, H.; Zhang, X.; Chen, X. miR-22 may suppress fibrogenesis by targeting TGFβR I in cardiac fibroblasts. Cell. Physiol. Biochem. 2016, 40, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Qi, Y.; Du, J.Q.; Zhang, D.F. MicroRNA-34a regulates cardiac fibrosis after myocardial infarction by targeting Smad4. Expert Opin. Ther. Targets 2014, 18, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Sun, X.; Shan, H.; Wang, N.; Wang, J.; Ren, J.; Feng, S.; Xie, L.; Lu, C.; Yuan, Y.; et al. miR-101 Inhibited Post-Infarct Cardiac Fibrosis and Improved Left Ventricular Compliance via FOS/TGFβ1 Pathway. Circulation 2012, 126, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Nguyen, S.S.; Gao, X.M.; Tham, Y.K.; Ooi, J.Y.; Patterson, N.L.; Kiriazis, H.; Su, Y.; Thomas, C.J.; Lin, R.C.; et al. Inhibition of miR-154 protects against cardiac dysfunction and fibrosis in a mouse model of pressure overload. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.B.; Raut, S.K.; Khanna, S.; Kumar, A.; Sharma, S.; Prasad, R.; Khullar, M. MicroRNA-200c modulates DUSP-1 expression in diabetes-induced cardiac hypertrophy. Mol. Cell. Biochem. 2017, 424, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Bei, Y.; Chen, P.; Lei, Z.; Fu, S.; Zhang, H.; Xu, J.; Che, L.; Chen, X.; Sluijter, J.P.; et al. Crucial role of miR-433 in regulating cardiac fibrosis. Theranostics 2016, 6, 2068–2083. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Donath, S.; Li, Y.; Qin, D.; Prabhakar, B.S.; Li, P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef]
- Wang, K.; An, T.; Zhou, L.Y.; Liu, C.Y.; Zhang, X.J.; Feng, C.; Li, P.F. E2F1-regulated miR-30b suppresses Cyclophilin D and protects heart from ischemia/reperfusion injury and necrotic cell death. Cell Death Differ. 2015, 22, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Sun, A.; Zhao, H.; Liu, X.; Zhang, W.; Jin, X.; Wang, C.; Ma, X.; Shen, C.; Zou, Y.; et al. MicroRNA-34a promotes cardiomyocyte apoptosis post myocardial infarction through down-regulating aldehyde dehydrogenase 2. Curr. Pharm. Des. 2013, 19, 4865–4873. [Google Scholar] [CrossRef]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Treguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Zhang, X.J.; Li, Q.; Wang, K.; Wang, Y.; Jiao, J.Q.; Feng, C.; Teng, S.; Zhou, L.Y.; Gong, Y.; et al. MicroRNA-103/107 regulate programmed necrosis and myocardial ischemia/reperfusion injury through targeting fadd. Circ. Res. 2015, 117, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Zhao, W.K.; Xu, S.J.; Fan, L.H.; Zhang, X.J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, D.L.; Long, B.; An, T.; Zhang, J.; Zhou, L.Y.; Liu, C.Y.; Li, P.F. NfAT4-dependent miR-324-5p regulates mitochondrial morphology and cardiomyocyte cell death by targeting Mtfr1. Cell Death Dis. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.Y.; Zhang, X.J.; Feng, C.; Zhou, L.Y.; Zhao, Y.; Li, P.F. miR-361-regulated prohibitin inhibits mitochondrial fission and apoptosis and protects heart from ischemia injury. Cell Death Differ. 2015, 22, 1058–1068. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Song, X.W.; Tian, J.; Chen, H.Y.; Li, D.F.; Wang, J.F.; Ren, A.J.; Yuan, W.J.; Lin, L. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 2012, 17, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhou, L.Y.; Wang, J.X.; Wang, Y.; Sun, T.; Zhao, B.; Yang, Y.J.; An, T.; Long, B.; Li, N.; et al. E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Jiao, J.Q.; Wang, J.X.; Liu, J.P.; Li, Q.; Li, P.F. miR-484 regulates mitochondrial network through targeting Fis1. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Jiao, J.Q.; Li, Q.; Long, B.; Wang, K.; Liu, J.P.; Li, Y.R.; Li, P.F. mR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 2011, 17, 71–78. [Google Scholar] [CrossRef]
- Wang, J.X.; Zhang, X.J.; Feng, C.; Sun, T.; Wang, K.; Wang, Y.; Zhou, L.Y.; Li, P.F. MicroRNA-532-3p regulates mitochondrial fission through targeting apoptosis repressor with caspase recruitment domain in doxorubicin cardiotoxicity. Cell Death Dis. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Zhou, L.Y.; Liu, F.; Zhou, Q.Y.; Liu, C.Y.; Fan, Y.Y.; Li, P.F. Carl lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Wang, K.; Li, N.; Murtaza, I.; Xiao, J.Y.; Fan, Y.Y.; Liu, C.Y.; Li, W.H.; Cheng, Z.; Li, P. miR-761 regulates the mitochondrial network by targeting mitochondrial fission factor. Free Radic. Biol. Med. 2013, 65, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Ding, S.L.; Long, B.; Liu, C.Y.; Sun, T.; Fan, Y.Y.; Sun, L.; Li, P.F. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Li, N.; Li, L.; Liu, C.Y.; Dong, Y.H.; Gao, J.N.; Zhou, L.Y.; Wang, C.Q.; Li, P.F. MicroRNA-2861 regulates programmed necrosis in cardiomyocyte by impairing adenine nucleotide translocase 1 expression. Free Radic. Biol. Med. 2016, 91, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhang, X.J.; Ji, Y.X.; Zhang, P.; Deng, K.Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhou, X.; Huang, J. Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.H.; Yuan, Y.X.; Rao, S.L.; Wang, P. LncRNA miat enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653–3660. [Google Scholar] [PubMed]
- Wang, K.; Liu, F.; Liu, C.Y.; An, T.; Zhang, J.; Zhou, L.Y.; Wang, M.; Dong, Y.H.; Li, N.; Gao, J.N.; et al. The long noncoding RNA NFR regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Q.; Wu, Y.B.; Yin, C.S.; Chen, L.; Zhang, Q.; Hu, L.Q. Obestatin attenuated doxorubicin-induced cardiomyopathy via enhancing long noncoding Mhrt RNA expression. Biomed. Pharmacother. 2016, 81, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sun, W.; Zhang, Z.; Zheng, Y. The role of Nrf2-mediated pathway in cardiac remodeling and heart failure. Oxid. Med. Cell. Longev. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, D.; Li, G.; Ming, X.; Tu, Y.; Tian, J.; Lu, H.; Yu, B. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell. Physiol. Biochem. 2015, 35, 1986–1998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sha, M.; Yao, Y.; Da, J.; Jing, D. Increased B-type-natriuretic peptide promotes myocardial cell apoptosis via the B-type-natriuretic peptide/long non-coding RNA LSINCT5/caspase-1/interleukin 1β signaling pathway. Mol. Med. Report. 2015, 12, 6761–6767. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.; Liang, J. Unraveling the expression profiles of long noncoding RNAs in rat cardiac hypertrophy and functions of lncRNA BC088254 in cardiac hypertrophy induced by transverse aortic constriction. Cardiology 2016, 134, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Zangrando, J.; Zhang, L.; Vausort, M.; Maskali, F.; Marie, P.Y.; Wagner, D.R.; Devaux, Y. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genom. 2014, 15, 460. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.P.; Ding, Y.; Chen, J.; Wu, G.; Kataoka, M.; Hu, Y.; Yang, J.H.; Liu, J.; Drakos, S.G.; Selzman, C.H.; et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc. Res. 2016, 112, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gu, H.; Xu, W.; Zhou, X. Down-regulation of lncRNA mMALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int. J. Cardiol. 2016, 203, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.; Hermans-Beijnsberger, S.; Beqqali, A.; Bitsch, N.; Nakagawa, S.; Prasanth, K.V.; de Windt, L.J.; van Oort, R.J.; Heymans, S.; Schroen, B. Long non-coding RNA Malat-1 is dispensable during pressure overload-induced cardiac remodeling and failure in mice. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Tan, W.L.; Lim, B.T.; Anene-Nzelu, C.G.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.; et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017, 113, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Jakobi, T.; Czaja-Hasse, L.F.; Reinhardt, R.; Dieterich, C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genom. Proteom. Bioinform. 2016, 14, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Werfel, S.; Nothjunge, S.; Schwarzmayr, T.; Strom, T.M.; Meitinger, T.; Engelhardt, S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016, 98, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.H.; Li, R.; Su, Y.M.; Xiao, J.; Pan, M.; Cai, X.X.; Ji, X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.M.; Zhang, M.; Huang, L.; Hu, Z.Q.; Zhu, J.N.; Xiao, Z.; Zhang, Z.; Lin, Q.X.; Zheng, X.L.; Yang, M.; et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.Z.; Yang, F.; Tan, W.; Li, X.; Jiao, C.; Huang, R.; Yang, B.B. The anti-cancer components of ganoderma lucidum possesses cardiovascular protective effect by regulating circular RNA expression. Oncoscience 2016, 3, 203–207. [Google Scholar] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Gao, J.; Ding, S.L.; Wang, K.; Jiao, J.Q.; Wang, Y.; Sun, T.; Zhou, L.Y.; Long, B.; Zhang, X.J.; et al. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Mol. Cell 2015, 59, 50–61. [Google Scholar] [CrossRef] [PubMed]

| ncRNAs | Length | Main Functions |
|---|---|---|
| tRNAs | 74~95 nt | Transfer amino acids to ribosomes during protein synthesis |
| rRNAs | 121~5000 nt | Ribosome components, directly involved in the synthesis of proteins in robosome |
| snRNAs | 100~300 nt | Process mRNA precursor (splicing and maturation) |
| snoRNAs | 100~300 nt | Guide chemical modifications of other RNAs, such as rRNAs, tRNAs and snRNAs |
| gRNAs | 55~70 nt | Participate in RNA editing |
| miRNAs | 19~23 nt | Negatively regulate gene expression by promote mRNA degradation or inhibit mRNA translation |
| piRNAs | 24~30 nt | Play roles in gametogenesis, maintain transposon silencing, translation suppression, epigenetic regulation |
| siRNAs | 21~25 nt | Silence complementary target mRNA |
| lncRNAs | >200 nt | Regulate gene expression in epigenetic, transcriptional, post-transcriptional levels, miRNA sponge |
| circRNAs | Circular | As competing endogenous RNA or miRNA sponge; regulate alternative splicing and parental gene expression |
| miRNAs | Effects | Target Genes/ Signaling Pathway | Pathological Conditions | Reference |
|---|---|---|---|---|
| Hypertrophy | ||||
| let-7a | Anti- | calmodulin | Ang II | [19] |
| miR-1 | Anti- | HADC4 | Thyroid hormone | [20] |
| miR-9 | Anti- | myocardin | Iso and Aldo | [21] |
| miR-10a | Anti- | Tbx5 | Ang II, TAC | [22] |
| miR-22 | Anti- | PURB, Sirt1, Hdac4 | Pressure overload | [23,24] |
| miR-23a | Pro- | MuRF1 | Iso and Aldo | [25] |
| miR-26a | Anti- | GATA4 | Ang II, TAC | [26] |
| miR-34 | Pro- | VEGF, vinculin, POFUT1, Notch1, SEMA4B | TAC | [27] |
| miR-101 | Anti- | Rab1a | Ang II, TAC | [28] |
| miR-155 | Pro- | Jarid2 | TAC, activated calcineurin Tg | [29] |
| miR-182 | Pro- | Bcat2, Foxo3, Adcy6/Akt | PIGF | [30] |
| miR-206 | Pro- | Foxp1 | YAP, pressure overload | [31] |
| miR-212/132 | Pro- | Foxo3 | Pressure overload | [32] |
| miR-223 | Pro- | ARC | Iso, TAC | [33] |
| miR-365 | Pro- | Skp2/ mTOR | Ang II | [34] |
| miR-378 | Anti- | MAPK1, IGFR1, GRB2, KSR1/MAPK | TAC | [35] |
| miR-489 | Anti- | MyD88 | Ang II, TAC | [36] |
| miR-497 | Anti- | Sirt4 | Ang II, TAC | [37] |
| miR-541 | Anti- | - | Ang II | [38] |
| Fibrosis | ||||
| miR-1 | Anti- | Fibullin-2/MAPK | Pressure overload | [39] |
| miR-21 | Pro- | Spry1/ERK-MAP kinase PTEN/SMAD7 | Pressure overload, Ang II | [40,41] |
| miR-22 | Anti- | TGFβRI | Ang II | [42] |
| miR-30 | Anti- | CTGF | Renin-2 Tg | [43] |
| miR-34a | Pro- | Smad4 | MI | [44] |
| miR-101 | Anti- | c-Fos/TGF-β1 | Ang II, CAL | [45] |
| miR-154 | Pro- | Atg7 | TAC | [46] |
| miR-200c | Pro- | DUSP-1/MAPK | High glucose | [47] |
| miR-433 | Pro- | AZIN1, JNK1/TGFβ/Smad3 | MI | [48] |
| Apoptosis/Necrosis | ||||
| miR-30 | Anti- | p53, Cyclophilin D | H2O2, I/R | [49,50] |
| miR-34a | Pro- | PPP1R10, ALDH2 | MI | [51,52] |
| miR-103/107 | Pro- | FADD | I/R, H2O2 | [53] |
| miR-188-3p | Anti- | Atg7 | I/R, A/R | [54] |
| miR-324-5p | Anti- | Mtfr1 | I/R, A/R | [55] |
| miR-361 | Pro- | PHB1 | M/I, H2O2 | [56] |
| miR-378 | Anti- | Caspase-3 | CAL/Hypoxia | [57] |
| miR-421 | Pro- | Pink1 | I/R, H2O2 | [58] |
| miR-484 | Anti- | Fission 1 | I/R, Anoxia, Dox | [59] |
| miR-499 | Anti- | CnAα, CnAβ | I/R, Anoxia | [60] |
| miR-532-3p | Pro- | ARC | Dox | [61] |
| miR-539 | Pro- | PHB2 | I/R, Anoxia | [62] |
| miR-761 | Anti- | MFF | I/R, H2O2 | [63] |
| miR-874 | Pro- | Caspase-8 | I/R, H2O2 | [64] |
| miR-2861 | Pro- | ANT1 | I/R, H2O2 | [65] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Xu, W.; Wang, J.; Wang, K.; Li, P. The Role and Molecular Mechanism of Non-Coding RNAs in Pathological Cardiac Remodeling. Int. J. Mol. Sci. 2017, 18, 608. https://doi.org/10.3390/ijms18030608
Gao J, Xu W, Wang J, Wang K, Li P. The Role and Molecular Mechanism of Non-Coding RNAs in Pathological Cardiac Remodeling. International Journal of Molecular Sciences. 2017; 18(3):608. https://doi.org/10.3390/ijms18030608
Chicago/Turabian StyleGao, Jinning, Wenhua Xu, Jianxun Wang, Kun Wang, and Peifeng Li. 2017. "The Role and Molecular Mechanism of Non-Coding RNAs in Pathological Cardiac Remodeling" International Journal of Molecular Sciences 18, no. 3: 608. https://doi.org/10.3390/ijms18030608





